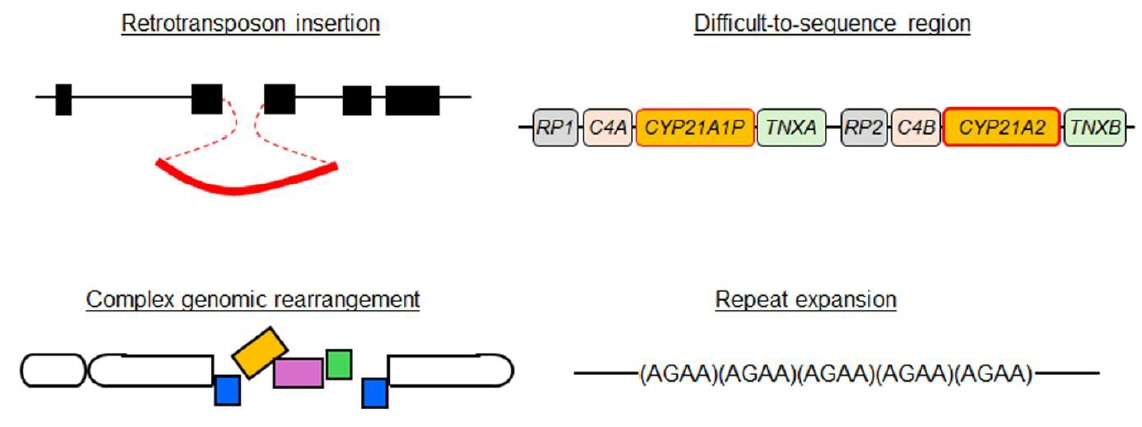
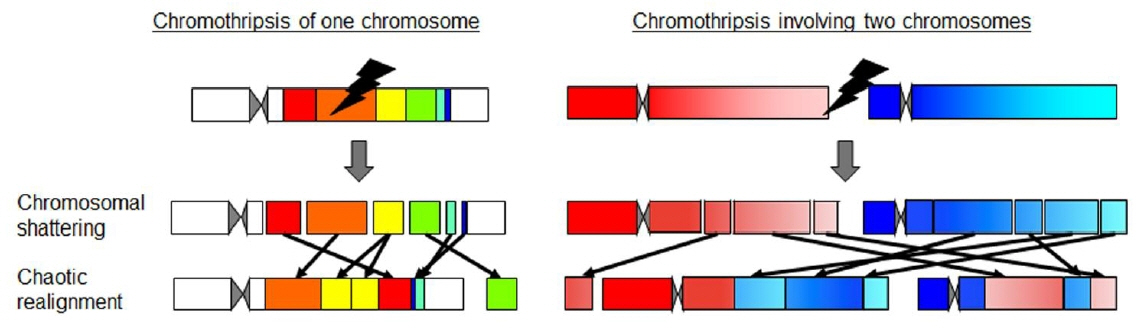
Ann Pediatr Endocrinol Metab.
2024 Jun;29(3):156-160. 10.6065/apem.2448028.014.
Long-read next-generation sequencing for molecular diagnosis of pediatric endocrine disorders
- Affiliations
-
- 1Division of Diversity Research, National Research Institute for Child Health and Development, Tokyo, Japan
- 2Department of Genome Medicine, National Research Institute for Child Health and Development, Tokyo, Japan
- 3Department of Molecular Endocrinology, National Research Institute for Child Health and Development, Tokyo, Japan
- KMID: 2556671
- DOI: http://doi.org/10.6065/apem.2448028.014
Abstract
- Recent advances in long-read next-generation sequencing (NGS) have enabled researchers to identify several pathogenic variants overlooked by short-read NGS, array-based comparative genomic hybridization, and other conventional methods. Long-read NGS is particularly useful in the detection of structural variants and repeat expansions. Furthermore, it can be used for mutation screening in difficultto- sequence regions, as well as for DNA-methylation analyses and haplotype phasing. This mini-review introduces the usefulness of long-read NGS in the molecular diagnosis of pediatric endocrine disorders.
Figure
Cited by 1 articles
-
Commentary on "Long-read next-generation sequencing for molecular diagnosis of pediatric endocrine disorders"
Won Kyoung Cho
Ann Pediatr Endocrinol Metab. 2024;29(3):141-141. doi: 10.6065/apem.24224014edi03.
Reference
-
References
1. Eichler EE. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med. 2019; 381:64–74.
Article2. Fukami M, Miyado M. Next generation sequencing and array-based comparative genomic hybridization for molecular diagnosis of pediatric endocrine disorders. Ann Pediatr Endocrinol Metab. 2017; 22:90–4.
Article3. Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007; 39(7 Suppl):S48–54.
Article4. Izumi Y, Suzuki E, Kanzaki S, Yatsuga S, Kinjo S, Igarashi M, et al. Genome-wide copy number analysis and systematic mutation screening in 58 patients with hypogonadotropic hypogonadism. Fertil Steril. 2014; 102:1130–6.
Article5. Suga K, Imoto I, Ito H, Naruto T, Goji A, Osumi K, et al. Next-generation sequencing for the diagnosis of patients with congenital multiple anomalies and/or intellectual disabilities. J Med Invest. 2020; 67:246–9.
Article6. Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020; 21:597–614.
Article7. Oehler JB, Wright H, Stark Z, Mallett AJ, Schmitz U. The application of long-read sequencing in clinical settings. Hum Genomics. 2023; 17:73.
Article8. Miller DE, Sulovari A, Wang T, Loucks H, Hoekzema K, Munson KM, et al. Targeted long-read sequencing identifies missing disease-causing variation. Am J Hum Genet. 2021; 108:1436–49.
Article9. Conlin LK, Aref-Eshghi E, McEldrew DA, Luo M, Rajagopalan R. Long-read sequencing for molecular diagnostics in constitutional genetic disorders. Hum Mutat. 2022; 43:1531–44.
Article10. Mastrorosa FK, Miller DE, Eichler EE. Applications of longread sequencing to Mendelian genetics. Genome Med. 2023; 15:42.
Article11. Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006; 7:407–42.
Article12. Stankiewicz P, Lupski JR. The genomic basis of disease, mechanisms and assays for genomic disorders. Genome Dyn. 2006; 1:1–16.
Article13. Miller DE, Hanna P, Galey M, Reyes M, Linglart A, Eichler EE, et al. Targeted long-read sequencing identifies a retrotransposon insertion as a cause of altered GNAS exon A/B methylation in a family with autosomal dominant pseudohypoparathyroidism type 1b (PHP1B). J Bone Miner Res. 2022; 37:1711–9.
Article14. Taşkesen M, Collin GB, Evsikov AV, Güzel A, Özgül RK, Marshall JD, et al. Novel Alu retrotransposon insertion leading to Alström syndrome. Hum Genet. 2012; 131:407–13.
Article15. Del Gobbo GF, Wang X, Couse M, Mackay L, Goldsmith C, Marshall AE, et al. Long-read genome sequencing reveals a novel intronic retroelement insertion in NR5A1 associated with 46,XY differences of sexual development. Am J Med Genet A. 2024; 194:e63522.
Article16. Pfaff AL, Singleton LM, Kõks S. Mechanisms of diseaseassociated SINE-VNTR-Alus. Exp Biol Med (Maywood). 2022; 247:756–64.17. Stevanovski I, Chintalaphani SR, Gamaarachchi H, Ferguson JM, Pineda SS, Scriba CK, et al. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci Adv. 2022; 8:e. abm5386.
Article18. Kekou K, Sofocleous C, Papadimas G, Petichakis D, Svingou M, Pons RM, et al. A dynamic trinucleotide repeat (TNR) expansion in the DMD gene. Mol Cell Probes. 2016; 30:254–60.
Article19. Miyatake S, Koshimizu E, Fujita A, Doi H, Okubo M, Wada T, et al. Rapid and comprehensive diagnostic method for repeat expansion diseases using nanopore sequencing. NPJ Genom Med. 2022; 7:62.
Article20. Liu P, Erez A, Nagamani SC, Dhar SU, Kołodziejska KE, Dharmadhikari AV, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011; 146:889–903.
Article21. Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011; 20:1916–24.
Article22. Hattori A, Fukami M. Established and novel mechanisms leading to de novo genomic rearrangements in the human germline. Cytogenet Genome Res. 2020; 160:167–76.
Article23. Lei M, Liang D, Yang Y, Mitsuhashi S, Katoh K, Miyake N, et al. Long-read DNA sequencing fully characterized chromothripsis in a patient with Langer-Giedion syndrome and Cornelia de Lange syndrome-4. J Hum Genet. 2020; 65:667–74.
Article24. Mantere T, Kersten S, Hoischen A. Long-read sequencing emerging in medical genetics. Front Genet. 2019; 10:426.
Article25. Stephens Z, Milosevic D, Kipp B, Grebe S, Iyer RK, Kocher JA. PB-Motif-A method for identifying gene/pseudogene rearrangements with long reads: an application to CYP21A2 genotyping. Front Genet. 2021; 12:716586.
Article26. Zhang R, Cui D, Song C, Ma X, Cai N, Zhang Y, et al. Evaluating the efficacy of a long-read sequencingbased approach in the clinical diagnosis of neonatal congenital adrenocortical hyperplasia. Clin Chim Acta. 2024; 555:117820.
Article27. Adachi E, Nakagawa R, Tsuji-Hosokawa A, Gau M, Kirino S, Yogi A, et al. A MinION-based long-read sequencing application with one-step PCR for the genetic diagnosis of 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2024; 109:750–60.
Article28. Soellner L, Begemann M, Mackay DJ, Grønskov K, Tümer Z, Maher ER, et al. Recent advances in imprinting disorders. Clin Genet. 2017; 91:3–13.
Article29. Yamada M, Okuno H, Okamoto N, Suzuki H, Miya F, Takenouchi T, et al. Diagnosis of Prader-Willi syndrome and Angelman syndrome by targeted nanopore long-read sequencing. Eur J Med Genet. 2023; 66:104690.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Commentary on "Long-read next-generation sequencing for molecular diagnosis of pediatric endocrine disorders"
- Next generation sequencing and array-based comparative genomic hybridization for molecular diagnosis of pediatric endocrine disorders
- Applications of genomic research in pediatric endocrine diseases
- Application of Next Generation Sequencing in Laboratory Medicine
- MinION(TM): New, Long Read, Portable Nucleic Acid Sequencing Device