J Vet Sci.
2014 Sep;15(3):389-398. 10.4142/jvs.2014.15.3.389.
Molecular characterization of duck enteritis virus CHv strain UL49.5 protein and its colocalization with glycoprotein M
- Affiliations
-
- 1Avian Disease Research Center, Sichuan Agricultural University, Chengdu 611130, China. jiary@sicau.edu.cn, chenganchun@vip.163.com
- 2Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu 611130, China.
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu 611130, China.
- KMID: 2155622
- DOI: http://doi.org/10.4142/jvs.2014.15.3.389
Abstract
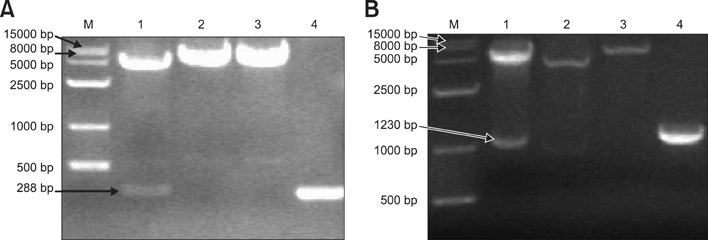
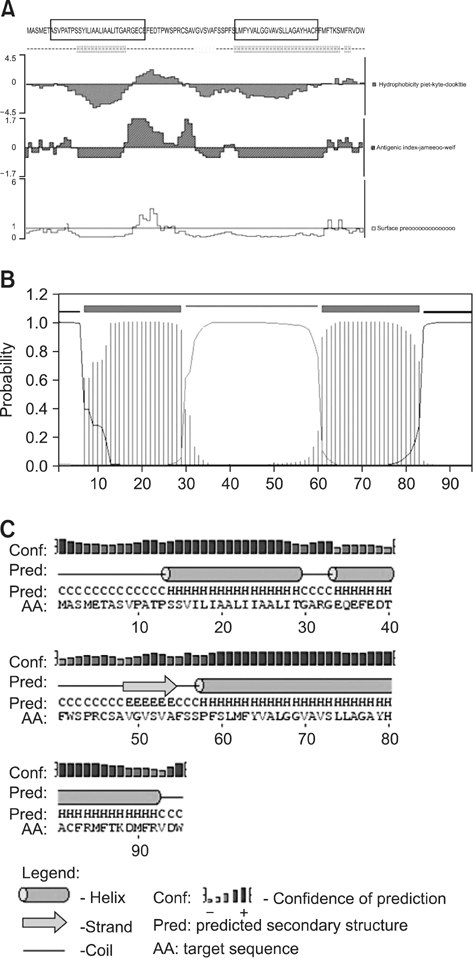
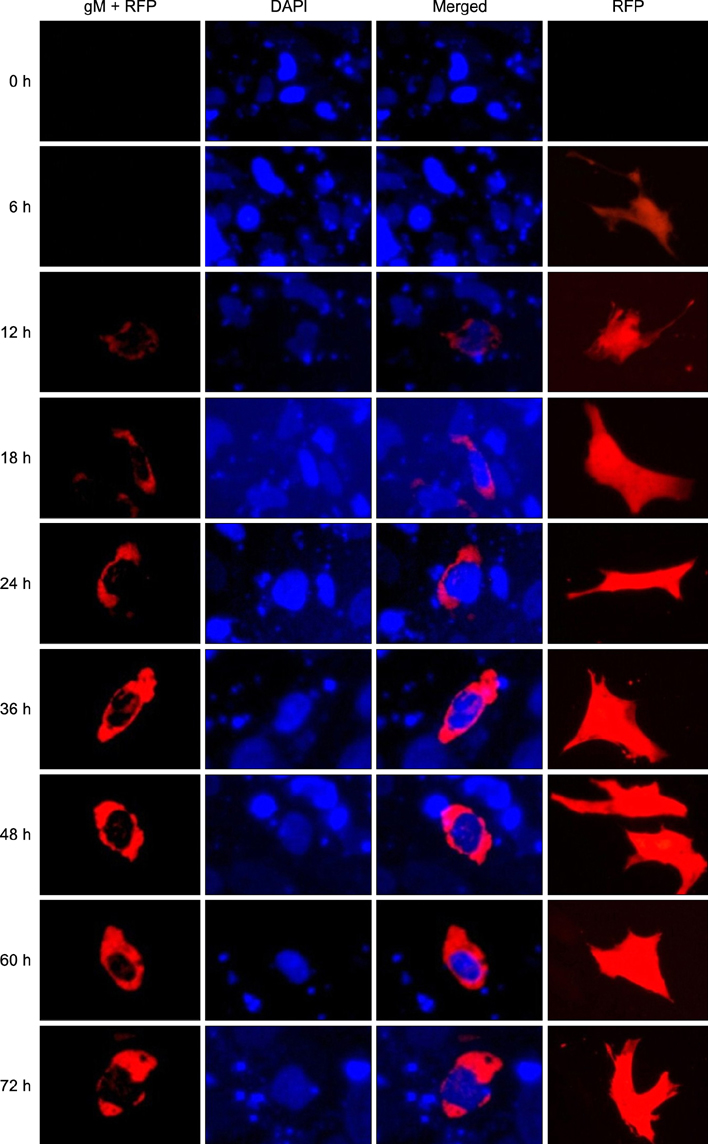
- The UL49.5 gene of most herpesviruses is conserved and encodes glycoprotein N. However, the UL49.5 protein of duck enteritis virus (DEV) (pUL49.5) has not been reported. In the current study, the DEV pUL49.5 gene was first subjected to molecular characterization. To verify the predicted intracellular localization of gene expression, the recombinant plasmid pEGFP-C1/pUL49.5 was constructed and used to transfect duck embryo fibroblasts. Next, the recombinant plasmid pDsRed1-N1/glycoprotein M (gM) was produced and used for co-transfection with the pEGFP-C1/pUL49.5 plasmid to determine whether DEV pUL49.5 and gM (a conserved protein in herpesviruses) colocalize. DEV pUL49.5 was thought to be an envelope glycoprotein with a signal peptide and two transmembrane domains. This protein was also predicted to localize in the cytoplasm and endoplasmic reticulum with a probability of 66.7%. Images taken by a fluorescence microscope at different time points revealed that the DEV pUL49.5 and gM proteins were both expressed in the cytoplasm. Overlap of the two different fluorescence signals appeared 12 h after transfection and continued to persist until the end of the experiment. These data indicate a possible interaction between DEV pUL49.5 and gM.
Keyword
MeSH Terms
Figure
Reference
-
1. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004; 340:783–795.
Article2. Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol. 2002; 20:83–87.
Article3. Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999; 294:1351–1362.
Article4. Burkhardt C, Himmelein S, Britt W, Winkler T, Mach M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J Gen Virol. 2009; 90:1951–1961.
Article5. Fuchs W, Mettenleiter TC. The nonessential UL49.5 gene of infectious laryngotracheitis virus encodes an O-glycosylated protein which forms a complex with the non-glycosylated UL10 gene product. Virus Res. 2005; 112:108–114.
Article6. Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000; 403:339–342.
Article7. Jöns A, Granzow H, Kuchling R, Mettenleiter TC. The UL49.5 gene of pseudorabies virus codes for an o-glycosylated structural protein of the viral envelope. J Virol. 1996; 70:1237–1241.
Article8. Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997; 5:147–152.9. Kirby AJ, Camilleri P, Engberts JBFN, Feiters MC, Nolte RJM, Söderman O, Bergsma M, Bell PC, Fielden ML, García Rodríguez CL, Guédat P, Kremer A, McGregor C, Perrin C, Ronsin G, van Eijk MCP. Gemini surfactants: new synthetic vectors for gene transfection. Angew Chem Int Ed Engl. 2003; 42:1448–1457.
Article10. Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J. 1998; 74:1591–1599.
Article11. Koppers-Lalic D, Reits EAJ, Ressing ME, Lipinska AD, Abele R, Koch J, Marcondes Rezende M, Admiraal P, van Leeuwen D, Bienkowska-Szewczyk K, Mettenleiter TC, Rijsewijk FAM, Tampé R, Neefjes J, Wiertz EJHJ. Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc Natl Acad Sci U S A. 2005; 102:5144–5149.
Article12. Koppers-Lalic D, Verweij MC, Lipińska AD, Wang Y, Quinten E, Reits EA, Koch J, Loch S, Marcondes Rezende M, Daus F, Bieńkowska-Szewczyk K, Osterrieder N, Mettenleiter TC, Heemskerk MHM, Tampé R, Neefjes JJ, Chowdhury SI, Ressing ME, Rijsewijk FAM, Wiertz EJHJ. Varicellovirus UL49.5 proteins differentially affect the function of the transporter associated with antigen processing, TAP. PLoS Pathog. 2008; 4:e1000080.
Article13. Koyano S, Mar EC, Stamey FR, Inoue N. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol. 2003; 84:1485–1491.
Article14. Kropff B, Burkhardt C, Schott J, Nentwich J, Fisch T, Britt W, Mach M. Glycoprotein N of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS Pathog. 2012; 8:e1002999.
Article15. Kullberg M, Mann K, Owens JL. A two-component drug delivery system using Her-2-targeting thermosensitive liposomes. J Drug Target. 2009; 17:98–107.
Article16. Li Y, Huang B, Ma X, Wu J, Li F, Ai W, Song M, Yang H. Molecular characterization of the genome of duck enteritis virus. Virology. 2009; 391:151–161.
Article17. Lipińska AD, Koppers-Lalic D, Rychłowski M, Admiraal P, Rijsewijk FAM, Bieńkowska-Szewczyk K, Wiertz EJHJ. Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J Virol. 2006; 80:5822–5832.
Article18. Mach M, Osinski K, Kropff B, Schloetzer-Schrehardt U, Krzyzaniak M, Britt W. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J Virol. 2007; 81:5212–5224.
Article19. March JC, Rao G, Bentley WE. Biotechnological applications of green fluorescent protein. Appl Microbiol Biotechnol. 2003; 62:303–315.
Article20. Masse MJ, Jöns A, Dijkstra JM, Mettenleiter TC, Flamand A. Glycoproteins gM and gN of pseudorabies virus are dispensable for viral penetration and propagation in the nervous systems of adult mice. J Virol. 1999; 73:10503–10507.
Article21. Mayhew TM, Griffiths G, Lucocq JM. Applications of an efficient method for comparing immunogold labelling patterns in the same sets of compartments in different groups of cells. Histochem Cell Biol. 2004; 122:171–177.
Article22. Nair R, Rost B. LOC3D: annotate sub-cellular localization for protein structures. Nucleic Acids Res. 2003; 31:3337–3340.
Article23. Pati SK, Novak Z, Purser M, Arora N, Mach M, Britt WJ, Boppana SB. Strain-specific neutralizing antibody responses against human cytomegalovirus envelope glycoprotein N. Clin Vaccine Immunol. 2012; 19:909–913.
Article24. Ren Y, Bell S, Zenner HL, Lau SYK, Crump CM. Glycoprotein M is important for the efficient incorporation of glycoprotein H-L into herpes simplex virus type 1 particles. J Gen Virol. 2012; 93:319–329.
Article25. Said A, Azab W, Damiani A, Osterrieder N. Equine herpesvirus type 4 UL56 and UL49.5 proteins downregulate cell surface major histocompatibility complex class I expression independently of each other. J Virol. 2012; 86:8059–8071.
Article26. Shimamura M, Mach M, Britt WJ. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol. 2006; 80:4591–4600.
Article27. Verheugt FW, von dem Borne AEG, Décary F, Engelfriet CP. The detection of granulocyte alloantibodies with an indirect immunofluorescence test. Br J Haematol. 1977; 36:533–544.
Article28. Wang C, Bian Z, Wei D, Zhang JG. miR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011; 352:197–207.
Article29. Xing J, Wang S, Li Y, Guo H, Zhao L, Pan W, Lin F, Zhu H, Wang L, Li M, Wang L, Zheng C. Characterization of the subcellular localization of herpes simplex virus type 1 proteins in living cells. Med Microbiol Immunol. 2011; 200:61–68.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Characterization of the L Segment of Hantann Virus, Strain Howang
- Molecular Characterization of a New Hantaan Virus Howang Strain
- Isolation and Molecular Characterization of Feline Herpesvirus 1 from Naturally Infected Korean Cats
- Immunocytochemical localization of myelin basic protein, proteolipid protein and myelin-associated glycoprotein in human oligodendrocyte in culture
- Molecular characterization of a 13-amino acid deletion in VP1 (1D) protein and novel amino acid substitutions in 3D polymerase protein of foot and mouth disease virus subtype A/Iran87