Immune Netw.
2014 Dec;14(6):321-327. 10.4110/in.2014.14.6.321.
SUMO Proteins are not Involved in TGF-beta1-induced, Smad3/4-mediated Germline alpha Transcription, but PIASy Suppresses it in CH12F3-2A B Cells
- Affiliations
-
- 1Department of Microbiology, College of Medicine, Konyang University, Daejeon 302-718, Korea. srpark@konyang.ac.kr
- 2Department of Molecular Bioscience, School of Biomedical Science, Kangwon National University, Chuncheon 200-701, Korea.
- 3Department of Biochemistry, College of Medicine, Konyang University, Daejeon 302-718, Korea.
- KMID: 2150822
- DOI: http://doi.org/10.4110/in.2014.14.6.321
Abstract
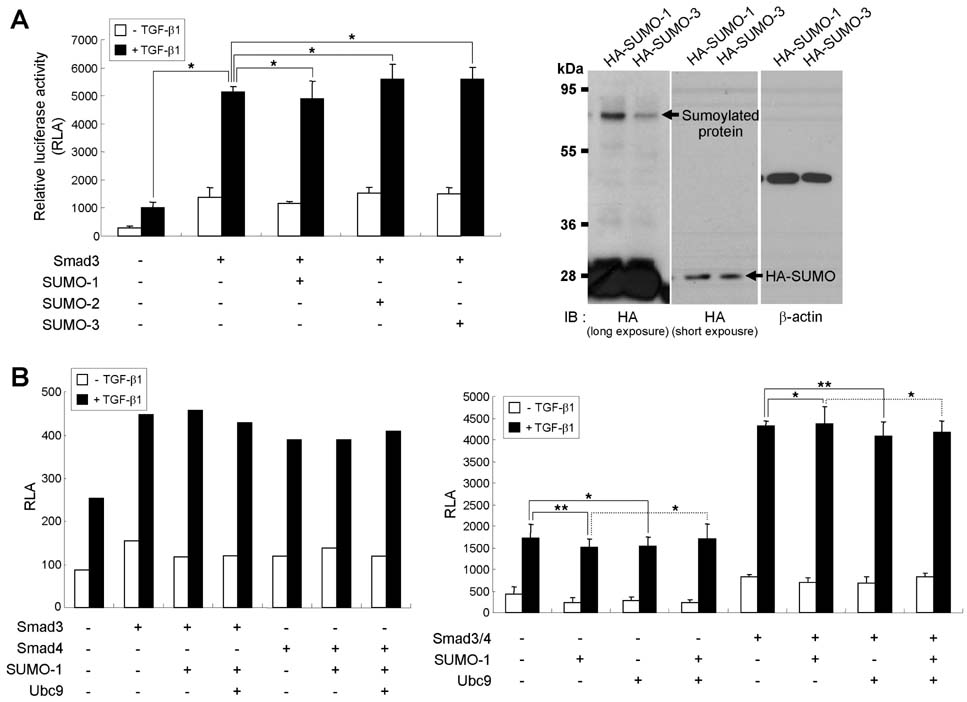
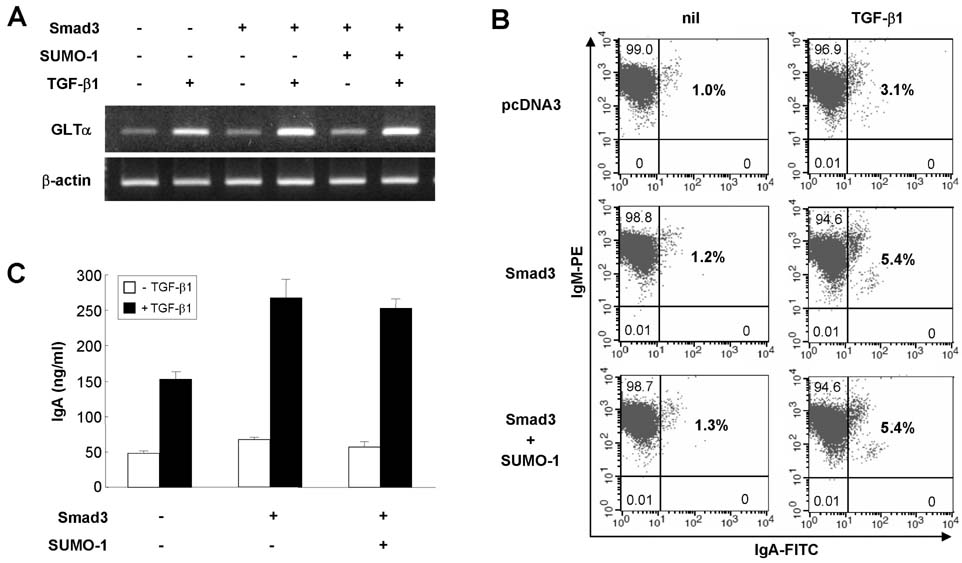
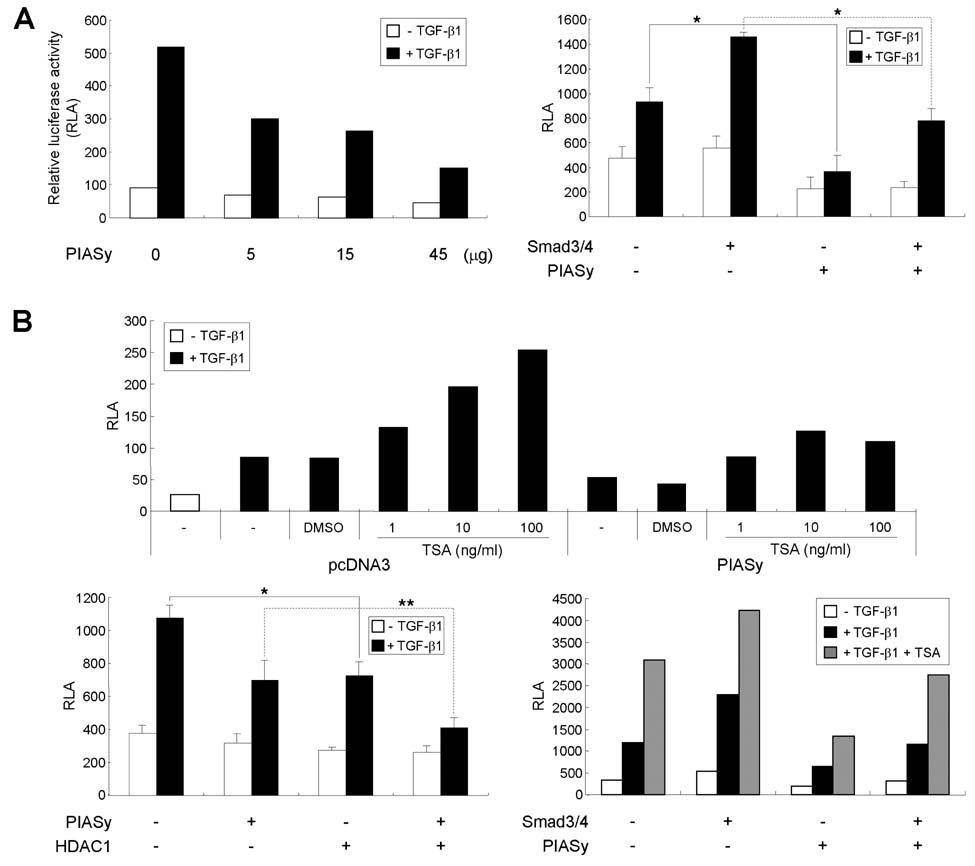
- TGF-beta induces IgA class switching by B cells. We previously reported that Smad3 and Smad4, pivotal TGF-beta signal-transducing transcription factors, mediate germline (GL) alpha transcription induced by TGF-beta1, resulting in IgA switching by mouse B cells. Post-translational sumoylation of Smad3 and Smad4 regulates TGF-beta-induced transcriptional activation in certain cell types. In the present study, we investigated the effect of sumoylation on TGF-beta1-induced, Smad3/4-mediated GLalpha transcription and IgA switching by mouse B cell line, CH12F3-2A. Overexpression of small ubiquitin-like modifier (SUMO)-1, SUMO-2 or SUMO-3 did not affect TGF-beta1-induced, Smad3/4-mediated GLalpha promoter activity, expression of endogenous GLalpha transcripts, surface IgA expression, and IgA production. Next, we tested the effect of the E3 ligase PIASy on TGF-beta1-induced, Smad3/4-mediated GLalpha promoter activity. We found that PIASy overexpression suppresses the GLalpha promoter activity in cooperation with histone deacetylase 1. Taken together, these results suggest that SUMO itself does not affect regulation of GLalpha transcription and IgA switching induced by TGF-beta1/Smad3/4, while PIASy acts as a repressor.
Keyword
MeSH Terms
-
Animals
B-Lymphocytes*
Cell Line
Histone Deacetylase 1
Immunoglobulin A
Immunoglobulin Class Switching
Mice
Small Ubiquitin-Related Modifier Proteins*
SUMO-1 Protein*
Sumoylation
Transcription Factors
Transcriptional Activation
Transforming Growth Factor beta
Transforming Growth Factor beta1
Ubiquitin-Protein Ligases
Histone Deacetylase 1
Immunoglobulin A
SUMO-1 Protein
Small Ubiquitin-Related Modifier Proteins
Transcription Factors
Transforming Growth Factor beta
Transforming Growth Factor beta1
Ubiquitin-Protein Ligases
Figure
Reference
-
1. Stavnezer J. Immunoglobulin class switching. Curr Opin Immunol. 1996; 8:199–205.
Article2. Stavnezer J. Molecular processes that regulate class switching. Curr Top Microbiol Immunol. 2000; 245:127–168.
Article3. Park SR. Activation-induced cytidine deaminase in B cell immunity and cancers. Immune Netw. 2012; 12:230–239.
Article4. Park SR, Lee JH, Kim PH. Smad3 and Smad4 mediate transforming growth factor-beta1-induced IgA expression in murine B lymphocytes. Eur J Immunol. 2001; 31:1706–1715.
Article5. Park SR, Lee EK, Kim BC, Kim PH. p300 cooperates with Smad3/4 and Runx3 in TGFbeta1-induced IgA isotype expression. Eur J Immunol. 2003; 33:3386–3392.
Article6. Choi SH, Seo GY, Nam EH, Jeon SH, Kim HA, Park JB, Kim PH. Opposing effects of Arkadia and Smurf on TGFbeta1-induced IgA isotype expression. Mol Cells. 2007; 24:283–287.7. Park KH, Nam EH, Seo GY, Seo SR, Kim PH. Tiul1 and TGIF are involved in downregulation of TGFbeta1-induced IgA isotype expression. Immune Netw. 2009; 9:248–254.
Article8. Park SR, Jung MH, Jeon SH, Park MH, Park KH, Lee MR, Kim PH. IFN-gamma down-regulates TGF-beta1-induced IgA expression through Stat1 and p300 signaling. Mol Cells. 2010; 29:57–62.
Article9. Miyazono K, Kamiya Y, Miyazawa K. SUMO amplifies TGF-beta signalling. Nat Cell Biol. 2008; 10:635–637.10. Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000; 16:591–626.
Article11. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005; 6:599–609.
Article12. Ohshima T, Shimotohno K. Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J Biol Chem. 2003; 278:50833–50842.
Article13. Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J Biol Chem. 2003; 278:31043–31048.
Article14. Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of transforming growth factor-beta signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J Biol Chem. 2003; 278:18714–18719.
Article15. Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem. 2003; 278:27853–27863.
Article16. Liang M, Melchior F, Feng XH, Lin X. Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J Biol Chem. 2004; 279:22857–22865.
Article17. Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T. Regulation of transforming growth factor-beta signaling by protein inhibitor of activated STAT, PIASy through Smad3. J Biol Chem. 2003; 278:34253–34258.
Article18. Imoto S, Ohbayashi N, Ikeda O, Kamitani S, Muromoto R, Sekine Y, Matsuda T. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem Biophys Res Commun. 2008; 370:359–365.
Article19. Long J, Matsuura I, He D, Wang G, Shuai K, Liu F. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc Natl Acad Sci U S A. 2003; 100:9791–9796.
Article20. Long J, Wang G, Matsuura I, He D, Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3). Proc Natl Acad Sci U S A. 2004; 101:99–104.
Article21. Kang JS, Saunier EF, Akhurst RJ, Derynck R. The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat Cell Biol. 2008; 10:654–664.
Article22. Nakamura M, Kondo S, Sugai M, Nazarea M, Imamura S, Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996; 8:193–201.
Article23. Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996; 383:168–172.
Article24. Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996; 271:350–353.
Article25. Long J, Wang G, He D, Liu F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem J. 2004; 379:23–29.
Article26. Tou L, Liu Q, Shivdasani RA. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol Cell Biol. 2004; 24:3132–3139.
Article27. Park MH, Park SR, Park MR, Kim YH, Kim PH. Retinoic acid induces expression of Ig germ line α transcript, an IgA isotype switching indicative, through retinoic acid receptor. Genes Genomics. 2011; 33:83–88.
Article28. Jang YS, Choi SH, Park SR, Kim HA, Park JB, Kim PH. Characterization of mouse B lymphoma cells (CH12F3-2A) for the study of IgA isotype switching. Immune Netw. 2004; 4:216–223.
Article29. Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003; 4:137–142.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characterization of Mouse B Lymphoma Cells (CH12F3-2A) for the Study of IgA Isotype Switching
- TGF-beta1 induces mouse dendritic cells to express VEGF and its receptor (Flt-1) under hypoxic conditions
- Cardamonin Suppresses TGF-beta1-Induced Epithelial Mesenchymal Transition via Restoring Protein Phosphatase 2A Expression
- Hypoxia-Inducible Factor 1α Regulates the Transforming Growth Factor β1/SMAD Family Member 3 Pathway to Promote Breast Cancer Progression
- Activin Receptor-Like Kinase 5 Inhibitor Attenuates Fibrosis in Fibroblasts Derived from Peyronie's Plaque