Clin Exp Otorhinolaryngol.
2015 Sep;8(3):211-217. 10.3342/ceo.2015.8.3.211.
Cordblood-Based High-Throughput Screening for Deafness Gene of 646 Newborns in Jinan Area of China
- Affiliations
-
- 1Department of Clinical Laboratory, Handan Central Hospital, Handan, China. sxdoccn@163.com
- KMID: 2117514
- DOI: http://doi.org/10.3342/ceo.2015.8.3.211
Abstract
OBJECTIVES
Infants with slight/mild or late-onset hearing impairment might be missed in universal newborn hearing screening (UNHS). We identified the mutation hot spot of common deaf gene in the newborns in Jinan area population by screening the mutation spot with neonate cord blood, in order to make clear whether the neonate cord blood for screening is feasible.
METHODS
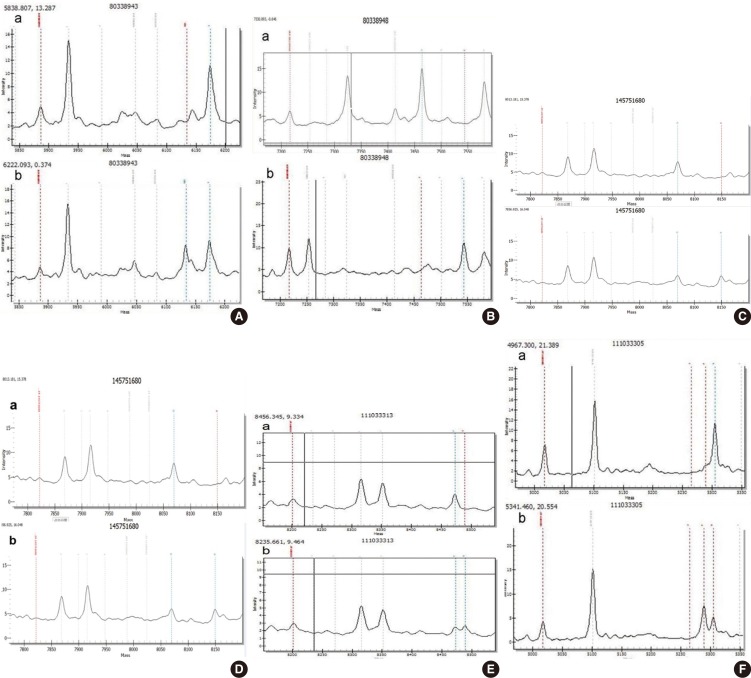
Six hundred and forty-six newborns were subjected to both UNHS and genetic screening for deafness by using neonate cord blood. The newborn genetic screening targeted four deafness-associated genes, which were commonly found in the Chinese population including gap junction beta-2 protein (GJB2), gap junction beta-3 protein (GJB3), solute carrier family 26 member 4 (SLC26A4), and mtDNA 12S rRNA. The most common 20 spot mutations in 4 deaf genes were detected by MassARRAY iPLEX platform and mitochondrial 12S rRNA A1555G and C1494T mutations were sequenced using Sanger sequencing.
RESULTS
Among the 646 newborns, 635 cases passed the UNHS and the other 11 cases (1.7%) did not. Of the 11 failures, two cases were found to carry homozygous GJB2 p.R143W pathogenic mutation, one case was found to have heterozygous GJB2 235delC mutation, and another one case carried heterozygous GJB3 p.R180X pathogenic mutation. Six hundred and thirty-five babies passed the newborn hearing screening, in which 25 babies were identified to carry pathogenic mutations, including 12 heterozygotes (1.9%) for GJB2 235delC, eight heterozygotes (1.3%) for SLC26A4 IVS7-2A>G, one heterozygote (0.2%) for p.R409H, two homozygotes (0.3%) for m.1494C>T, and two homozygotes (0.3%) for m.1555A>G.
CONCLUSION
Newborn genetic screening through the umbilical cord blood for common deafness-associated mutations may identify carriers sensitive to aminoglycoside antibiotic, and can effectively prevent or delay hearing loss occurs.
MeSH Terms
Figure
Reference
-
1. Ouyang XM, Yan D, Yuan HJ, Pu D, Du LL, Han DY, et al. The genetic bases for non-syndromic hearing loss among Chinese. J Hum Genet. 2009; 3. 54(3):131–140. PMID: 19197336.
Article2. Propst EJ, Stockley TL, Gordon KA, Harrison RV, Papsin BC. Ethnicity and mutations in GJB2 (connexin 26) and GJB6 (connexin 30) in a multi-cultural Canadian paediatric Cochlear Implant Program. Int J Pediatr Otorhinolaryngol. 2006; 3. 70(3):435–444. PMID: 16125251.
Article3. Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005; 12. 77(6):945–957. PMID: 16380907.4. Wang QJ, Zhao YL, Rao SQ, Guo YF, Yuan H, Zong L, et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin Genet. 2007; 9. 72(3):245–254. PMID: 17718863.
Article5. Kokotas H, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet. 2007; 5. 71(5):379–391. PMID: 17489842.
Article6. Kennedy C, McCann D. Universal neonatal hearing screening moving from evidence to practice. Arch Dis Child Fetal Neonatal Ed. 2004; 9. 89(5):F378–F383. PMID: 15321952.
Article7. Johnson JL, White KR, Widen JE, Gravel JS, James M, Kennalley T, et al. A multicenter evaluation of how many infants with permanent hearing loss pass a two-stage otoacoustic emissions/automated auditory brainstem response newborn hearing screening protocol. Pediatrics. 2005; 9. 116(3):663–672. PMID: 16140706.
Article8. Yoshinaga-Itano C, Sedey AL, Coulter DK, Mehl AL. Language of early- and later-identified children with hearing loss. Pediatrics. 1998; 11. 102(5):1161–1171. PMID: 9794949.
Article9. Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N Engl J Med. 1998; 2. 338(8):548–550. PMID: 9471561.
Article10. Li L, Lu J, Tao Z, Huang Q, Chai Y, Li X, et al. The p.V37I exclusive genotype of GJB2: a genetic risk-indicator of postnatal permanent childhood hearing impairment. PLoS One. 2012; 7(5):e36621. PMID: 22574200.
Article11. Oh SK, Choi SY, Yu SH, Lee KY, Hong JH, Hur SW, et al. Evaluation of the pathogenicity of GJB3 and GJB6 variants associated with nonsyndromic hearing loss. Biochim Biophys Acta. 2013; 1. 1832(1):285–291. PMID: 22617145.
Article12. Jackler RK, De La Cruz A. The large vestibular aqueduct syndrome. Laryngoscope. 1989; 12. 99(12):1238–1242. PMID: 2601537.
Article13. Reardon W, OMahoney CF, Trembath R, Jan H, Phelps PD. Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM. 2000; 2. 93(2):99–104. PMID: 10700480.
Article14. Pryor SP, Madeo AC, Reynolds JC, Sarlis NJ, Arnos KS, Nance WE, et al. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet. 2005; 2. 42(2):159–165. PMID: 15689455.
Article15. Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010; 6. 10(4):380–390. PMID: 20100600.
Article16. Nivoloni Kde A, da Silva-Costa SM, Pomílio MC, Pereira T, Lopes Kde C, de Moraes VC, et al. Newborn hearing screening and genetic testing in 8974 Brazilian neonates. Int J Pediatr Otorhinolaryngol. 2010; 8. 74(8):926–929. PMID: 20538352.17. Wu CC, Hung CC, Lin SY, Hsieh WS, Tsao PN, Lee CN, et al. Newborn genetic screening for hearing impairment: a preliminary study at a tertiary center. PLoS One. 2011; 6(7):e22314. PMID: 21811586.
Article18. Guan MX. Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Mitochondrion. 2011; 3. 11(2):237–245. PMID: 21047563.
Article19. Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004; 1. 74(1):139–152. PMID: 14681830.
Article20. Lu J, Qian Y, Li Z, Yang A, Zhu Y, Li R, et al. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion. 2010; 1. 10(1):69–81. PMID: 19818876.21. Schimmenti LA, Martinez A, Telatar M, Lai CH, Shapiro N, Fox M, et al. Infant hearing loss and connexin testing in a diverse population. Genet Med. 2008; 7. 10(7):517–524. PMID: 18580690.
Article22. Norris VW, Arnos KS, Hanks WD, Xia X, Nance WE, Pandya A. Does universal newborn hearing screening identify all children with GJB2 (Connexin 26) deafness? Penetrance of GJB2 deafness. Ear Hear. 2006; 12. 27(6):732–741. PMID: 17086082.
Article23. Young NM, Reilly BK, Burke L. Limitations of universal newborn hearing screening in early identification of pediatric cochlear implant candidates. Arch Otolaryngol Head Neck Surg. 2011; 3. 137(3):230–234. PMID: 21422305.
Article24. Pollak A, Skorka A, Mueller-Malesińska M, Kostrzewa G, Kisiel B, Waligora J, et al. M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A. 2007; 11. 143A(21):2534–2543. PMID: 17935238.25. Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003; 4. 40(4):242–248. PMID: 12676893.
Article26. Miyagawa M, Nishio SY, Usami S. Deafness Gene Study Consortium. Mutation spectrum and genotype-phenotype correlation of hearing loss patients caused by SLC26A4 mutations in the Japanese: a large cohort study. J Hum Genet. 2014; 5. 59(5):262–268. PMID: 24599119.
Article27. Choi BY, Stewart AK, Madeo AC, Pryor SP, Lenhard S, Kittles R, et al. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms? Hum Mutat. 2009; 4. 30(4):599–608. PMID: 19204907.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Onychocytic Matricoma: Report of an Asian Case
- Successful Treatment of Hailey-Hailey Disease with Aminolevulinic Acid Photodynamic Therapy
- Tissue-Engineered 3D In Vitro Disease Models for HighThroughput Drug Screening
- Single-Cell Landscape and a Macrophage Subset Enhancing Brown Adipocyte Function in Diabetes (Diabetes Metab J 2024;48:885-900)
- History and Practice of Thyroid Fine-Needle Aspiration in China, Based on Retrospective Study of the Practice in Shandong University Qilu Hospital