Ann Lab Med.
2015 Jul;35(4):423-428. 10.3343/alm.2015.35.4.423.
Comparison of FcRgamma-Deficient and CD57+ Natural Killer Cells Between Cord Blood and Adult Blood in the Cytomegalovirus-Endemic Korean Population
- Affiliations
-
- 1Department of Pediatrics, Chonnam National University Hwasun Hospital, Chonnam National University Medical School, Gwangju, Korea. hoonkook@chonnam.ac.kr
- 2Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, Korea. hasomii@hanmail.net
- 3Department of Laboratory Medicine, KS Hospital, Gwangju, Korea.
- 4Center for Creative Biomedical Scientists, Chonnam National University, Gwangju, Korea.
- 5Research Center for Cancer Immunotherapy, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 6Department of Anesthesiology and Pain Medicine, Chonnan National University Medical School and Hospital, Gwangju, Korea.
- 7Department of Companion & Laboratory Animal Science, Kongju National University, Yesan, Korea.
- 8Department of Biochemistry, Microbiology and Immunology, University of Ottawa, Ottawa, Canada.
- KMID: 2045865
- DOI: http://doi.org/10.3343/alm.2015.35.4.423
Abstract
- BACKGROUND
FcRgamma-deficient natural killer (NK) cells (g-NK cells) have been associated with cytomegalovirus (CMV) infection. However, the frequency of g-NK cells in a CMV-endemic area (i.e., Korea) has not yet been studied. We examined the frequency of g-NK cells and expression of CD57 on NK cells in cord blood (CB) and adult blood (AB).
METHODS
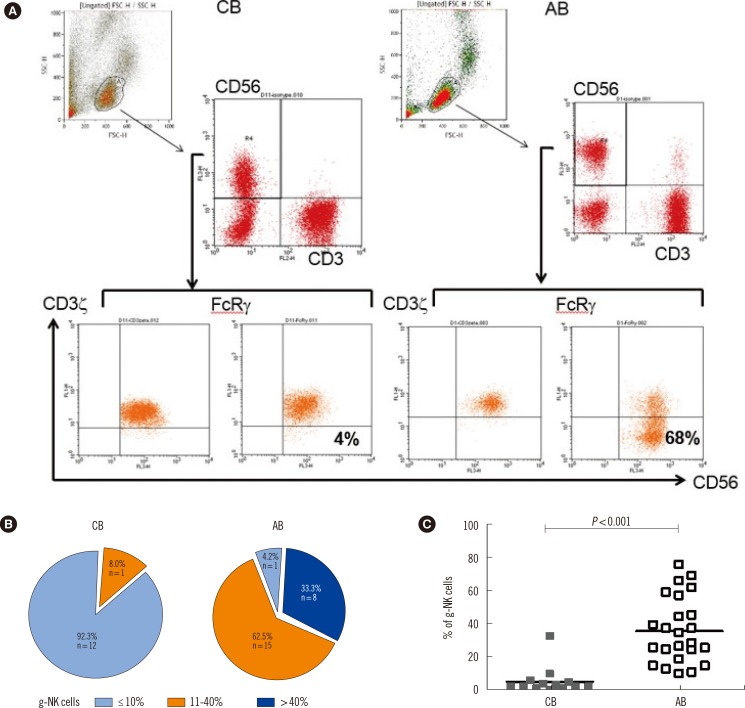
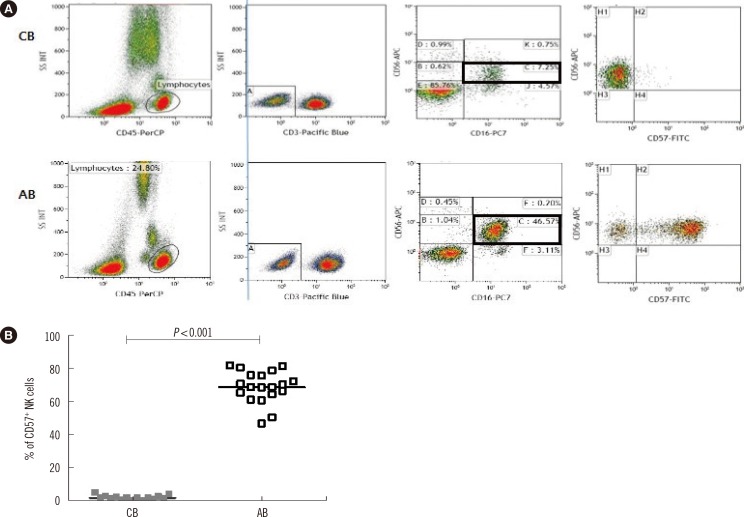
Of the 24 AB samples collected, 95.8% (23/24) were CMV IgG+/IgM-, while 100% of the 13 healthy CB samples were CMV IgG+/IgM-. We performed whole-blood flow cytometry assays to analyze intracellular FcRgamma and CD3zeta expression of CD3-/CD56dim NK cells from 13 CB and 24 AB samples, and surface CD57 expression on CD3-/CD56dim/CD16+ NK cells from 13 CB and 19 AB samples.
RESULTS
All CMV seropositive AB samples contained g-NK cells (23/23), and the median proportion of g-NK cells in the CD3-/CD56dim NK cell pool was 35.0% (range: 11-77%). CD57+ NK cells in the CD3-/CD56dim/CD16+ NK cell population were detected in all 19 AB samples tested, but not in any CB samples.
CONCLUSIONS
Our data suggest that g-NK cells and CD57+ NK cells are present at a very high frequency in CMV-seropositive AB, but rare in CMV-naive CB.
Keyword
Figure
Reference
-
1. Muntasell A, Vilches C, Angulo A, López-Botet M. Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host pathogen interaction. Eur J Immunol. 2013; 43:1133–1141. PMID: 23552990.2. Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003; 15:308–314. PMID: 12787756.
Article3. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008; 9:495–502. PMID: 18425106.
Article4. Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T, et al. Identification of human NK cells that are deficient for signaling adaptor FcRɣ and specialized for antibody-dependent immune functions. Int Immunol. 2012; 24:793–802. PMID: 22962434.
Article5. Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRɣ deficiency. J Immunol. 2013; 190:1402–1406. PMID: 23345329.
Article6. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012; 119:2665–2674. PMID: 22180440.
Article7. Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006; 43:1143–1151. PMID: 17029132.
Article8. Lopo S, Vinagre E, Palminha P, Paixao MT, Nogueira P, Freitas MG. Seroprevalence to cytomegalovirus in the Portuguese population, 2002-2003. Euro Surveill. 2011; 16:pii:19896.
Article9. Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010; 20:202–213. PMID: 20564615.
Article10. Sohn YM, Park KI, Lee C, Han DG, Lee WY. Congenital cytomegalovirus infection in Korean population with very high prevalence of maternal immunity. J Korean Med Sci. 1992; 7:47–51. PMID: 1329845.
Article11. Naitou H, Mimaya J, Horikoshi Y, Takashima Y, Amano K, Morita T. Qualitative and quantitative detection of cytomegalovirus DNA in sera by PCR as a clinical marker. Biol Pharm Bull. 1998; 21:1371–1375. PMID: 9881658.
Article12. Gaytant MA, Steegers EA, Semmekrot BA, Merkus HM, Galama JM. Congenital cytomegalovirus infection: review of the epidemiology and outcome. Obstet Gynecol Surv. 2002; 57:245–256. PMID: 11961482.
Article13. Tamul KR, Schmitz JL, Kane K, Folds JD. Comparison of the effects of Ficoll-Hypaque separation and whole blood lysis on results of immunophenotypic analysis of blood and bone marrow samples from patients with hematologic malignancies. Clin Diagn Lab Immunol. 1995; 2:337–342. PMID: 7545079.
Article14. Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2011; 108:14725–14732. PMID: 21825173.15. Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011; 208:13–21. PMID: 21173105.
Article16. Marcenaro E, Carlomagno S, Pesce S, Della Chiesa M, Parolini S, Moretta A, et al. NK cells and their receptors during viral infections. Immunotherapy. 2011; 3:1075–1086. PMID: 21913830.
Article17. Gumá M, Budt M, Sáez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006; 107:3624–3631. PMID: 16384928.18. Gumá M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, et al. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006; 194:38–41. PMID: 16741880.
Article19. Della Chiesa M, Marcenaro E, Sivori S, Carlomagno S, Pesce S, Moretta A. Human NK cell response to pathogens. Semin Immunol. 2014; 26:152–160. PMID: 24582551.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Phenotypic Analysis of Lymphocyte Subpopulation in Cord Blood
- Association of CD57+ Natural Killer Cells with Better Overall Survival in DLBCL Patients
- The Prognostic Significance of Intratumoral Natural Killer Cells in Colorectal Cancer
- A Study of Natural Killer Cell Activities , T Cells and T Cell Subsets in Vitiligo
- CD57 (Leu-7, HNK-1) immunoreactivity seen in thin arteries in the human fetal lung