Tuberc Respir Dis.
2008 Apr;64(4):285-292. 10.4046/trd.2008.64.4.285.
Diagnosis of Malignant Pleural Effusion by using Aberrant Methylation of p16 and RARB2
- Affiliations
-
- 1Department of Internal Medicine, Dong-A University College of Medicine, Busan, Korea. son30243@hanmail.net
- 2Department of Pathology, Dong-A University College of Medicine, Busan, Korea.
- 3Department of Radiology, Dong-A University College of Medicine, Busan, Korea.
- 4Department of Thoracic and Cardiovascular Surgery, Dong-A University College of Medicine, Busan, Korea.
- KMID: 1970179
- DOI: http://doi.org/10.4046/trd.2008.64.4.285
Abstract
-
BACKGROUND: A diagnosis of malignant pleural effusion is clinically important, as the prognosis of lung cancer patients with malignant pleural effusion is poor. The diagnosis will be difficult if a cytological test is negative. This study was performed to investigate whether the detection of hypermethylation of the p16 (CDKN2A) and retinoic acid receptor b2 (RARB2) genes in pleural fluid is useful for a diagnosis of malignant pleural effusion.
METHODS
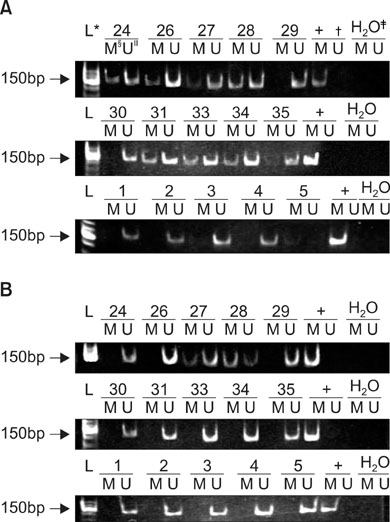
Pleural effusion was collected from 43 patients and was investigated for the aberrant promoter methylation of the RARB2 and CDKN2A genes by use of methylation-specific PCR. Results were compared with findings from a pleural biopsy and from pleural fluid cytology.
RESULTS
Of 43 cases, 17 cases of pleural effusion were due to benign diseases, and 26 cases were from lung cancer patients with malignant pleural effusion. Hypermethylation of the RARB2 and CDKN2A genes was not detected in the case of benign diseases, independent of whether or not the patients had ever smoked. In 26 cases of malignant pleural effusion, hypermethylation of RARB2, CDKN2A or either of these genes was detected in 14, 5 and 15 cases, respectively. The sensitivities of a pleural biopsy, pleural fluid cytology, hypermethylation of RARB2, hypermethylation of CDKN2A, or hypermethylation of either of the genes were 73.1%, 53.8%, 53.8%, 19.2%, and 57.7%, respectively; negative predictive values were 70.8%, 58.6%, 58.6%, 44.7%, and 60.7%, respectively. If both genes are considered together, the sensitivity and negative predictive value was lower than that for a pleural biopsy, but higher than that for pleural fluid cytology. The sensitivity of hypermethylation of the RARB2 gene for malignant pleural effusion was lower in small cell lung cancers than in non-small cell lung cancers.
CONCLUSION
These results demonstrate that detection of hypermethylation of the RARB2 and CDKN2A genes showed a high specificity, and sensitivity was higher than for pleural fluid cytology. With a better understanding of the pathogenesis of lung cancer according to histological types at the molecular level, and if appropriate genes are selected for hypermethylation testing, more precise results may be obtained.
Keyword
MeSH Terms
Figure
Reference
-
1. Annual report on the cause of death statistics, 2005. Korea National Statistical Office. 2005. Daejeon, Korea: Korea National Statistical Office;available from: http://www.cancer.go.kr/nciapps/user/basicinfo/gcancer_main.jsp.2. Light RW. Clinical practice. Pleural effusion. N Engl J Med. 2002. 346:1971–1977.3. Poe RH, Israel RH, Utell MJ, Hall WJ, Greenblatt DW, Kallay MC. Sensitivity, specificity, and predictive values of closed pleural biopsy. Arch Intern Med. 1984. 144:325–328.4. Digel W, Lubbert M. DNA methylation disturbances as novel therapeutic target in lung cancer: preclinical and clinical results. Crit Rev Oncol Hematol. 2005. 55:1–11.5. Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. Aberrant methylation of P16INK4a is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998. 95:11891–11896.6. Houle B, Rochette-Egly C, Bradley WE. Tumor-suppressive effect of the retinoic acid receptor beta in human epidermoid lung cancer cells. Proc Natl Acad Sci U S A. 1993. 90:985–989.7. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996. 93:9821–9826.8. Schmiemann V, Böcking A, Kazimirek M, Onofre AS, Gabbert HE, Kappes R, et al. Methylation assay for the diagnosis of lung cancer on bronchial aspirates: a cohort study. Clin Cancer Res. 2005. 11:7728–7734.9. Kim YC, Kwon YS, Oh IJ, Kim KS, Kim SY, Ryu JS, et al. National survey of lung cancer in Korea, 2005. J Lung Cancer. 2007. 6:67–73.10. Postmus PE, Brambilla E, Chansky K, Crowley J, Goldstraw P, Patz EF Jr, et al. The IASLC lung cancer staging project: proposals for revision of the M descriptors in the forthcoming (seventh) edition of the TNM classification of lung cancer. J Thorac Oncol. 2007. 2:686–693.11. Gilliland FD, Harms HJ, Crowell RE, Li Y-F, Willink R, Belinsky SA. Glutathione s-transferase P1 and NADPH quinone oxidoreductase polymorphisms are associated with aberrant promoter methylation of P16INK4a and O6-methylguanine-DNA methyltransferase in sputum. Cancer Res. 2002. 62:2248–2252.12. Fraipont F, Moro-Sibilot D, Michelland S, Brambilla E, Brambilla C, Favrot MC. Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer. 2005. 50:199–209.13. Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, Edelson MI, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004. 64:6476–6481.14. Liu Y, Lee MO, Wang HG, Li Y, Hashimoto Y, Klaus M, et al. Retinoic acid receptor beta mediates the growth inhibitory effect of retinoic acid by promoting apoptosis in human breast cancer cells. Mol Cell Biol. 1996. 16:1138–1149.15. Cote S, Sinnett D, Momparler RL. Demethylation by 5-aza-2'-deoxycytidine of specific 5-methylcytosine sites in the promoter region of the retinoic acid receptor beta gene in human colon carcinoma cells. Anticancer Drugs. 1998. 9:743–750.16. Katayama H, Hiraki A, Aoe K, Fujiwara K, Matsuo K, Maeda T, et al. Aberrant promoter methylation in pleural fluid DNA for diagnosis of malignant pleural effusion. Int J Cancer. 2007. 120:2191–2195.17. Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer cell lines that retains wildtype RB. Oncogene. 1994. 9:3375–3378.18. Toyooka S, Toyooka KO, Maruyama R, Virmani AK, Girard L, Miyajima K, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001. 1:61–67.19. Grote HJ, Schmiemann V, Geddert H, Rohr UP, Kappes R, Gabbert HE, et al. Aberrant promoter methylation of p16INK4a, RARB2 and SEMA3B in bronchial aspirates from patients with suspected lung cancer. Int J Cancer. 2005. 116:720–725.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Methylation of p16 and E-cadherin in ameloblastoma
- The Utility of Pleural Adenosine Deaminase for Diagnosis of Differentiating Tuberculous Pleural Effusion in Children
- Diagnostic Value of LDH and its Isoenzyme in Pleural Effusion
- Malignant Pleural Effusion: Medical Approaches for Diagnosis and Management
- Diagnostic Value of Adenosine Deaminase(ADA) and its Isoenzyme in Pleural Effusion