Cancer Res Treat.
2005 Aug;37(4):233-240.
Gene Promoter Hypermethylation in Tumors and Plasma of Breast Cancer Patients
- Affiliations
-
- 1Department of Pathology, Yeungnam University College of Medicine, Daegu, Korea. ykbae@yumail.ac.kr
- 2Department of Surgery, Yeungnam University College of Medicine, Daegu, Korea.
- 3Department of Preventive Medicine and Public Health, Yeungnam University College of Medicine, Daegu, Korea.
- 4Department of Radiation Oncology, Yeungnam University College of Medicine, Daegu, Korea.
- 5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Abstract
- PURPOSE
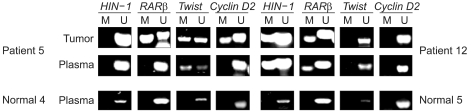
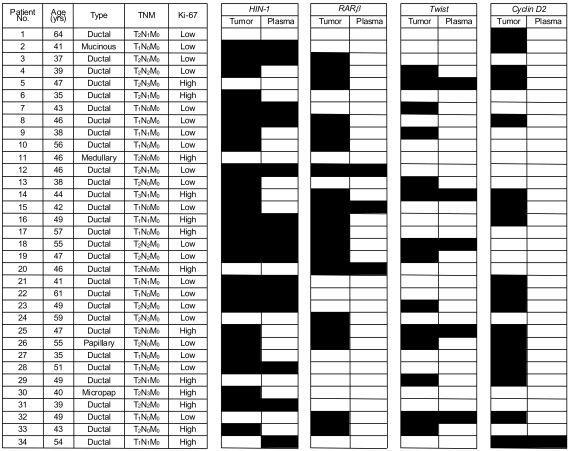
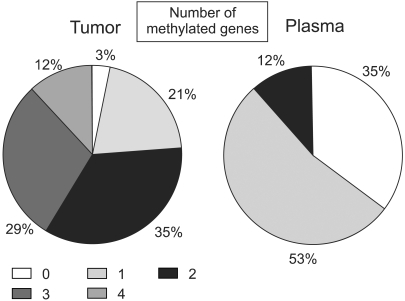
To measure the hypermethylation of four genes in primary tumors and paired plasma samples to determine the feasibility of gene promoter hypermethylation markers for detecting breast cancer in the plasma. MATERIALS AND METHODS: DNA was extracted from the tumor tissues and peripheral blood plasma of 34 patients with invasive breast cancer, and the samples examined for aberrant hypermethylation in cyclin D2, retinoic acid receptor beta (RARbeta), twist and high in normal-1 (HIN-1) genes using methylation-specific PCR (MSP), and the results correlated with the clinicopathological parameters. RESULTS: Promoter hypermethylation was detected at high frequency in the primary tumors for cyclin D2 (53%), RARbeta (56%), twist (41%) and HIN-1 (77%). Thirty-three of the 34 (97%) primary tumors displayed promoter hypermethylation in at least one of the genes examined. The corresponding plasma samples showed hyperme thylation of the same genes, although at lower frequencies (6% for cyclin D2, 16% for RARbeta, 36% for twist, and 54% for HIN-1). Overall, 22 of the 33 (67%) primary tumors with hypermethylation of at least one of the four genes also had abnormally hypermethylated DNA in their matched plasma samples. No significant relationship was recognized between any of the clinical or pathological parameters (tumor size, axillary lymph node metastasis, stage, or Ki-67 labeling index) with the frequency of hypermethylated DNA in the primary tumor or plasma. CONCLUSION: The detection of aberrant promoter hypermethylation of cancer-related genes in the plasma may be a useful tool for the detection of breast cancer.
Keyword
MeSH Terms
Figure
Reference
-
1. Shin HR, Jung KW, Won YJ, Park JG. 139 KCCR-affiliated Hospitals. 2002 annual report of the Korea central cancer registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004; 36:103–114.
Article2. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61:1659–1665. PMID: 11245480.3. Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996; 2:1035–1037. PMID: 8782464.
Article4. Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996; 2:1033–1035. PMID: 8782463.
Article5. Hibi K, Robinson CR, Booker S, Wu L, Hamilton SR, Sidransky D, et al. Molecular detection of genetic alterations in serum of colorectal cancer patients. Cancer Res. 1998; 58:1405–1407. PMID: 9537240.6. Mulcahy HE, Lyautey J, Lederrey C, qi Chen X, Anker P, Alstead EM, et al. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res. 1998; 4:271–275. PMID: 9516910.7. Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999; 59:67–70. PMID: 9892187.8. Sanchez-cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, et al. Gene promoter hypermehtylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000; 60:892–895. PMID: 10706101.9. Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999; 59:71–73. PMID: 9892188.10. Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001; 61:900–902. PMID: 11221878.
Article11. Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, et al. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002; 8:1761–1766. PMID: 12060614.12. Silva JM, Dominguez G, Garcia JM, Gonzalez R, Villanueva MJ, Navarro F, et al. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res. 1999; 59:3251–3256. PMID: 10397273.13. Muller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, et al. DNA methylation in serum of breast cancer patients: An independent prognostic marker. Cancer Res. 2003; 63:7641–7645. PMID: 14633683.14. Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004; 10:6189–6193. PMID: 15448006.
Article15. Hu XC, Wong IH, Chow LW. Tumor-derived aberrant methylation in plasma of invasive ductal breast cancer patients: clinical implications. Oncol Rep. 2003; 10:1811–1815. PMID: 14534701.
Article16. Bae YK, Brown A, Garrett E, Bornman D, Fackler MJ, Sukumar S, et al. Hypermethylation in histologically distinct classes of breast cancer. Clin Cancer Res. 2004; 10:5998–6005. PMID: 15447983.
Article17. Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, et al. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001; 61:2782–2787. PMID: 11289162.18. Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, Polyak K, et al. DNA methylation of RASSF1A, HIN-1, RAR-β, cyclin D2 and twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003; 107:970–975. PMID: 14601057.19. Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, Seth P, et al. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci USA. 2001; 98:9796–9801. PMID: 11481438.
Article20. Anbazhagan R, Fujii H, Gabrielson E. Microsatellite instability is uncommon in breast cancer. Clin Cancer Res. 1999; 5:839–844. PMID: 10213220.21. Borresen-Dale AL. TP53 and breast cancer. Hum Mutat. 2003; 21:292–300. PMID: 12619115.22. Cottrell S, Laird RW. Sensitive detection of DNA methylation. Ann N Y Acad Sci. 2003; 983:120–130. PMID: 12724217.
Article23. Evron E, Dooley WC, Umbricht CB, Rosenthal D, Sacchi N, Gabrielson E, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001; 357:1335–1336. PMID: 11343741.
Article24. Taback B, Hoon DS. Circulating nucleic acids and proteomics of plasma/serum: clinical utility. Ann N Y Acad Sci. 2004; 1022:1–8. PMID: 15251932.
Article25. Fujiwara K, Fujimoto N, Tabata M, Nishii K, Matsuo K, Hotta K, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res. 2005; 11:1219–1225. PMID: 15709192.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Downregulation of the RUNX3 Gene by Promoter Hypermethylation and Hemizygous Deletion in Breast Cancer
- The Role of the CDH1 Promoter Hypermethylation in the Axillary Lymph Node Metastasis and Prognosis
- Loss of PTEN Expression in Breast Cancers
- Promoter hypermethylation and loss of heterozygosity of FHIT genes in squamous cell carcinoma of uterine cervix
- Comparison of Methylation Profiling in Cancerous and Their Corresponding Normal Tissues from Korean Patients with Breast Cancer