Ann Lab Med.
2014 May;34(3):203-209. 10.3343/alm.2014.34.3.203.
Evaluation of Propidium Monoazide Real-Time PCR for Early Detection of Viable Mycobacterium tuberculosis in Clinical Respiratory Specimens
- Affiliations
-
- 1Department of Laboratory Medicine, Pusan National University School of Medicine, Yangsan, Korea. cchl@pusan.ac.kr
- 2Department of Physical Medicine & Rehabilitation, Korea University College of Medicine, Seoul, Korea.
- 3Department of Social Studies of Medicine, Pusan National University School of Medicine, Yangsan, Korea.
- 4Research Institute for Convergence of Biomedical Science and Technology, Pusan National University Yangsan Hospital, Yangsan, Korea.
- KMID: 1791921
- DOI: http://doi.org/10.3343/alm.2014.34.3.203
Abstract
- BACKGROUND
Conventional acid-fast bacilli (AFB) staining cannot differentiate viable from dead cells. Propidium monoazide (PMA) is a photoreactive DNA-binding dye that inhibits PCR amplification by DNA modification. We evaluated whether PMA real-time PCR is suitable for the early detection of viable Mycobacterium tuberculosis (MTB) in clinical respiratory specimens.
METHODS
A total of 15 diluted suspensions from 5 clinical MTB isolates were quadruplicated and subjected to PMA treatment and/or heat inactivation. Eighty-three AFB-positive sputum samples were also tested to compare the DeltaC(T) values (C(T) value in PMA-treated sputum samples-C(T) value in non-PMA-treated sputum samples) between culture-positive and culture-negative specimens. Real-time PCR was performed using Anyplex MTB/NTM Real-Time Detection (Seegene, Korea), and the C(T) value changes after PMA treatment were compared between culture-positive and culture-negative groups.
RESULTS
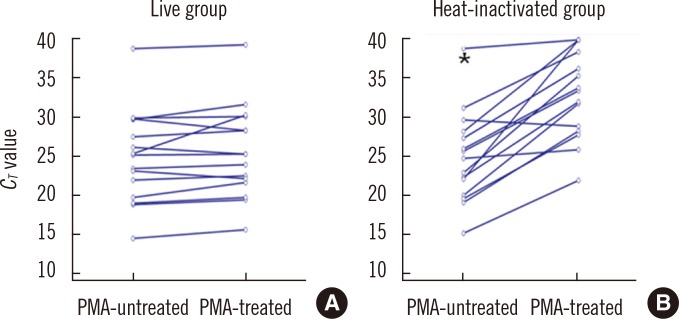
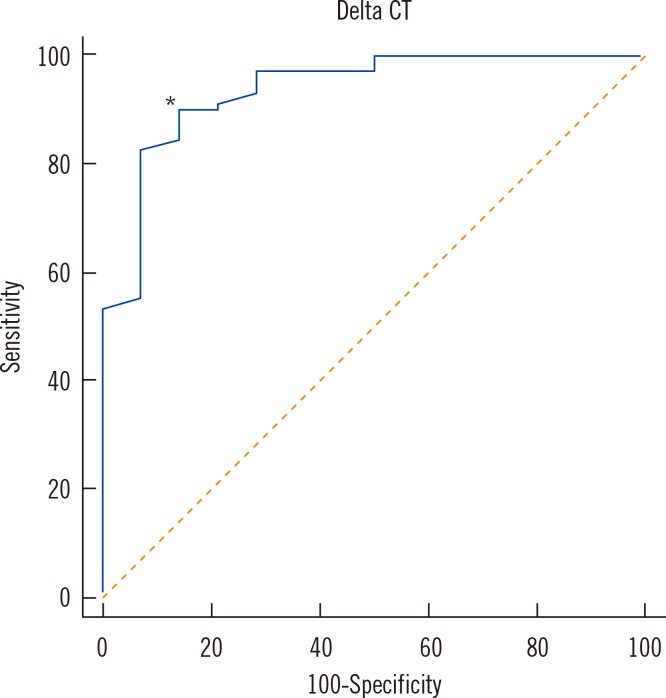
In MTB suspensions, the increase in the C(T) value after PMA treatment was significant in dead cells (P=0.0001) but not in live cells (P=0.1070). In 14 culture-negative sputum samples, the median DeltaC(T) value was 5.3 (95% confidence interval [CI], 4.1-8.2; P<0.0001), whereas that in 69 culture-positive sputum samples was 1.1 (95% CI, 0.7-2.0). In the ROC curve analysis, the cutoff DeltaC(T) value for maximum sensitivity (89.9%) and specificity (85.7%) for differentiating dead from live cells was 3.4.
CONCLUSIONS
PMA real-time PCR is a useful approach for differentiating dead from live bacilli in AFB smear-positive sputum samples.
MeSH Terms
-
Adult
Aged
Area Under Curve
Azides/*chemistry
DNA, Bacterial/*analysis
Female
Humans
Lung Diseases/diagnosis/*microbiology/pathology
Male
Middle Aged
Mycobacterium tuberculosis/genetics/*isolation & purification
Pilot Projects
Propidium/*analogs & derivatives/chemistry
ROC Curve
*Real-Time Polymerase Chain Reaction
Sputum/microbiology
Tuberculosis/*diagnosis/microbiology
Azides
DNA, Bacterial
Propidium
Figure
Cited by 1 articles
-
Effect of temperature and medium on the viability of Mycobacterium leprae during long term-storage
Jin-Ho Park, Yun-Ji Kim, Jong-Pill Kim
Korean Lepr Bull. 2019;52(1):41-50. doi: 10.33161/klb.2019.52.1.41.
Reference
-
1. Drobniewski FA, Caws M, Gibson A, Young D. Modern laboratory diagnosis of tuberculosis. Lancet Infect Dis. 2003; 3:141–147. PMID: 12614730.
Article2. Mishra A, Singhal A, Chauhan DS, Katoch VM, Srivastava K, Thakral SS, et al. Direct detection and identification of Mycobacterium tuberculosis and Mycobacterium bovis in bovine samples by a novel nested PCR assay: correlation with conventional techniques. J Clin Microbiol. 2005; 43:5670–5678. PMID: 16272503.3. Su WJ. Recent advances in the molecular diagnosis of tuberculosis. J Microbiol Immunol Infect. 2002; 35:209–214. PMID: 12542245.4. Chang HE, Heo SR, Yoo KC, Song SH, Kim SH, Kim HB, et al. Detection of Mycobacterium tuberculosis complex using real-time polymerase chain reaction. Korean J Lab Med. 2008; 28:103–108. PMID: 18458505.
Article5. Causse M, Ruiz P, Gutiérrez-Aroca JB, Casal M. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol. 2011; 49:3065–3067. PMID: 21653775.
Article6. Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med. 2003; 167:603–662. PMID: 12588714.7. Fittipaldi M, Nocker A, Codony F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J Microbiol Methods. 2012; 91:276–289. PMID: 22940102.
Article8. World Health Organization. Laboratory services in tuberculosis control. Part II. Microscopy. WHO/TB/98.258. Geneva, Switzerland: World Health Organization;1998.9. van der Kuyp F, Mahan CS. Prolonged positivity of sputum smears with negative cultures during treatment for pulmonary tuberculosis. Int J Tuberc Lung Dis. 2012; 16:1663–1667. PMID: 23131266.
Article10. Brescia CC, Griffin SM, Ware MW, Varughese EA, Egorov AI, Villegas EN. Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl Environ Microbiol. 2009; 75:6856–6863. PMID: 19749067.11. Yáñez MA, Nocker A, Soria-Soria E, Múrtula R, Martínez L, Catalán V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J Microbiol Methods. 2011; 85:124–130. PMID: 21329735.
Article12. Nocker A, Mazza A, Masson L, Camper AK, Brousseau R. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J Microbiol Methods. 2009; 76:253–261. PMID: 19103234.
Article13. Miotto P, Bigoni S, Migliori GB, Matteelli A, Cirillo DM. Early tuberculosis treatment monitoring by Xpert(R) MTB/RIF. Eur Respir J. 2012; 39:1269–1271. PMID: 22547737.14. Nkuipou-Kenfack E, Engel H, Fakih S, Nocker A. Improving efficiency of viability-PCR for selective detection of live cells. J Microbiol Methods. 2013; 93:20–24. PMID: 23389080.
Article15. Pholwat S, Heysell S, Stroup S, Foongladda S, Houpt E. Rapid first- and second-line drug susceptibility assay for Mycobacterium tuberculosis isolates by use of quantitative PCR. J Clin Microbiol. 2011; 49:69–75. PMID: 21084506.
Article16. Kang H, Sung N, Lee S, Kim D, Jeon D, Hwang S, et al. Comparison of smear and culture positivity using NaOH method and NALC-NaOH method for sputum treatment. Tuberc Respir Dis. 2008; 65:379–384.
Article17. Pan Y. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl Environ Microbiol. 2007; 73:8028–8031. PMID: 17933922.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of Propidium Monoazide Real-Time PCR for Viablity of Mycobacterium leprae
- Selective detection of viable Enterococcus faecalis using propidium monoazide in combination with real-time PCR
- Effect of temperature and medium on the viability of Mycobacterium leprae during long term-storage
- Evaluation of the Performances of AdvanSure TB/NTM Real Time PCR Kit for Detection of Mycobacteria in Respiratory Specimens
- Clinical Usefulness of Real-time PCR and Amplicor MTB PCR Assays for Diagnosis of Tuberculosis