J Korean Med Sci.
2012 Oct;27(10):1155-1161. 10.3346/jkms.2012.27.10.1155.
Rapid Detection of Prognostically Significant Fusion Transcripts in Acute Leukemia Using Simplified Multiplex Reverse Transcription Polymerase Chain Reaction
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea. hschi@amc.seoul.kr
- KMID: 1778820
- DOI: http://doi.org/10.3346/jkms.2012.27.10.1155
Abstract
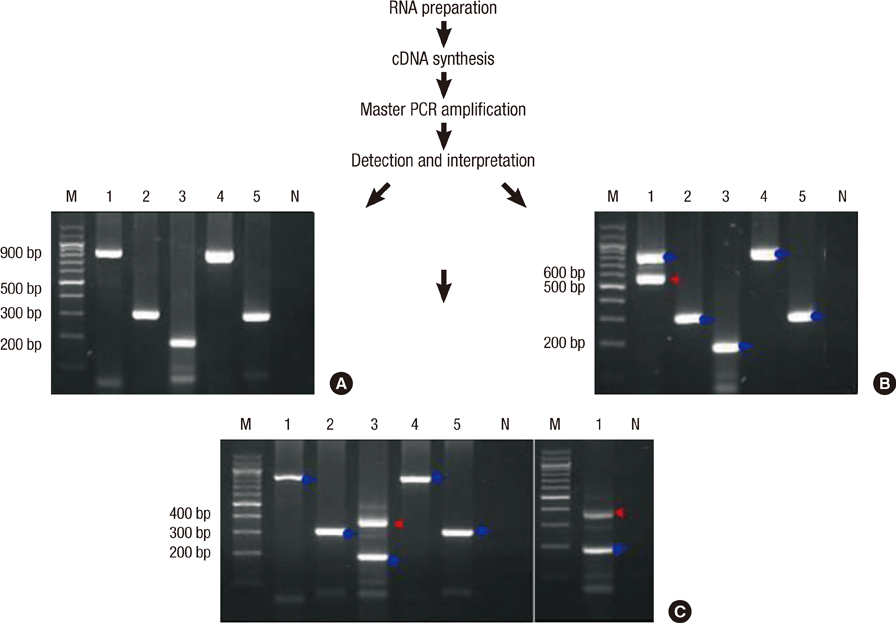
- Multiplex reverse transcription polymerase chain reaction (mRT-PCR) has recently emerged as an alternative to cytogenetics. We designed and used simplified mRT-PCR system as a molecular screen for acute leukemia. Fifteen fusion transcripts were included: BCR-ABL1, PML-RARA, ZBTB16-RARA, RUNX1-RUNX1T1, CBFB-MYH11, DEK-NUP214, TCF3-PBX1, ETV6-RUNX1, MLL-AFF1, MLL-MLLT4, MLL-MLLT3, MLL-MLLT10, MLL-ELL, MLL-MLLT1, and MLL-MLLT6. A total of 121 diagnostic acute leukemia specimens were studied, comparing the mRT-PCR system with standard cytogenetics. Fifty-six cases (46.3%) had fusion transcripts revealed by our mRT-PCR assay. The concordance rate between mRT-PCR and cytogenetics was 91.7%. However, false negative results were found in three cases who have inv(16), t(4;11) or t(11;19)(q23;p13.1), respectively. Seven cryptic translocations including ETV6-RUNX1, MLL-MLLT3, MLL-MLLT4, and PML-RARA were detected. This mRT-PCR assay is a useful screening tool in acute leukemia because it provides rapid and reliable detection of clinically important chimeric transcripts. In addition, cryptic translocations provide additional genetic information that could be clinically useful.
MeSH Terms
Figure
Reference
-
1. Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004. 18:115–136.2. Borkhardt A, Cazzaniga G, Viehmann S, Valsecchi MG, Ludwig WD, Burci L, Mangioni S, Schrapppe M, Riehm H, Lampert F, et al. Associazione Italiana Ematologia Oncologia Pediatrica and the Berlin-Frankfurt-Münster Study Group. Incidence and clinical relevance of TEL/AML1 fusion genes in children with acute lymphoblastic leukemia enrolled in the German and Italian multicenter therapy trials. Blood. 1997. 90:571–577.3. Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M, Hovi L, LeBlanc T, Szczepanski T, Ferster A, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007. 370:240–250.4. Dombret H, Preudhomme C, Boissel N. Core binding factor acute myeloid leukemia (CBF-AML): is high-dose Ara-C (HDAC) consolidation as effective as you think? Curr Opin Hematol. 2009. 16:92–97.5. Yanada M, Ohno R, Naoe T. Recent advances in the treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Int J Hematol. 2009. 89:3–13.6. Thomas X, Raffoux E, Renneville A, Pautas C, de Botton S, Terre C, Gardin C, Hayette S, Preudhomme C, Dombret H. Which AML subsets benefit from leukemic cell priming during chemotherapy? Long-term analysis of the ALFA-9802 GM-CSF study. Cancer. 2010. 116:1725–1732.7. Hunger SP, Devaraj PE, Foroni L, Secker-Walker LM, Cleary ML. Two types of genomic rearrangements create alternative E2A-HLF fusion proteins in t(17;19)-ALL. Blood. 1994. 83:2970–2977.8. Uphoff CC, MacLeod RA, Denkmann SA, Golub TR, Borkhardt A, Janssen JW, Drexler HG. Occurrence of TEL-AML1 fusion resulting from (12;21) translocation in human early B-lineage leukemia cell lines. Leukemia. 1997. 11:441–447.9. Uckun FM, Herman-Hatten K, Crotty ML, Sensel MG, Sather HN, Tuel-Ahlgren L, Sarquis MB, Bostrom B, Nachman JB, Steinherz PG, et al. Clinical significance of MLL-AF4 fusion transcript expression in the absence of a cytogenetically detectable t(4;11)(q21;q23) chromosomal translocation. Blood. 1998. 92:810–821.10. Grimwade D, Biondi A, Mozziconacci MJ, Hagemeijer A, Berger R, Neat M, Howe K, Dastugue N, Jansen J, Radford-Weiss I, et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): results of the European Working Party. Blood. 2000. 96:1297–1308.11. O'Reilly J, Chipper L, Springall F, Herrmann R. A unique structural abnormality of chromosome 16 resulting in a CBF beta-MYH11 fusion transcript in a patient with acute myeloid leukemia, FAB M4. Cancer Genet Cytogenet. 2000. 121:52–55.12. Rowe D, Cotterill SJ, Ross FM, Bunyan DJ, Vickers SJ, Byron J, McMullan DJ, Griffiths MJ, Reilly JT, Vandenberghe EA, et al. Cytogenetically cryptic AML1-ETO and CBF beta-MYH11 gene rearrangements: incidence in 412 cases of acute myeloid leukemia. Br J Haematol. 2000. 111:1051–1056.13. von Bergh A, Gargallo P, De Prijck B, Vranckx H, Marschalek R, Larripa I, Kluin P, Schuuring E, Hagemeijer A. Cryptic t(4;11) encoding MLL-AF4 due to insertion of 5' MLL sequences in chromosome 4. Leukemia. 2001. 15:595–600.14. Brown J, Jawad M, Twigg SR, Saracoglu K, Sauerbrey A, Thomas AE, Elis R, Harbott J, Kearney L. A cryptic t(5;11)(q35;p15.5) in 2 children with acute myeloid leukemia with apparently normal karyotypes, identified by a multiplex fluorescence in situ hybridization telomere assay. Blood. 2002. 99:2526–2531.15. Helias C, Leymarie V, Entz-Werle N, Falkenrodt A, Eyer D, Costa JA, Cherif D, Lutz P, Lessard M. Translocation t(5;14)(q35;q32) in three cases of childhood T cell acute lymphoblastic leukemia: a new recurring and cryptic abnormality. Leukemia. 2002. 16:7–12.16. Han JY, Kim KE, Kim KH, Park JI, Kim JS. Identification of PML-RARA rearrangement by RT-PCR and sequencing in an acute promyelocytic leukemia without t(15;17) on G-banding and FISH. Leuk Res. 2007. 31:239–243.17. Strehl S, König M, Mann G, Haas OA. Multiplex reverse transcriptase-polymerase chain reaction screening in childhood acute myeloblastic leukemia. Blood. 2001. 97:805–808.18. Elia L, Mancini M, Moleti L, Meloni G, Buffolino S, Krampera M, De Rossi G, Foa R, Cimino G. A multiplex reverse transcription-polymerase chain reaction strategy for the diagnostic molecular screening of chimeric genes: a clinical evaluation on 170 patients with acute lymphoblastic leukemia. Haematologica. 2003. 88:275–279.19. Olesen LH, Clausen N, Dimitrijevic A, Kerndrup G, Kjeldsen E, Hokland P. Prospective application of a multiplex reverse transcription-polymerase chain reaction assay for the detection of balanced translocation in leukaemia: a single-laboratory study of 390 paediatric and adult patients. Br J Haematol. 2004. 127:59–66.20. Hutchings Hoffmann M, Wirenfeldt Klausen T, Hasle H, Schmiegelow K, Brondum-Nielsen K, Johnsen HE. Multiplex reverse transcription polymerase chain reaction screening in acute myeloid leukemia detects cytogenetically unrevealed abnormalities of prognostic significance. Haematologica. 2005. 90:984–986.21. Preiss BS, Kerndrup GB, Pedersen RK, Hasle H, Pallisgaard N. Contribution of multiparameter genetic analysis to the detection of genetic alterations in hematologic neoplasia. An evaluation of combining G-band analysis, spectral karyotyping, and multiplex reverse-transcription polymerase chain reaction (multiplex RT-PCR). Cancer Genet Cytogenet. 2006. 165:1–8.22. Meyer-Monard S, Parlier V, Passweg J, Muhlematter D, Hess U, Bargetzi M, Kuhne T, Cabrol C, Gratwohl A, Jotterand M, et al. Combination of broad molecular screening and cytogenetic analysis for genetic risk assignment and diagnosis in patients with acute leukemia. Leukemia. 2006. 20:247–253.23. Park JS, Yi JW, Jeong SH, Lee HW, Kang SY, Choi JH, Kim HC, Park JE, Kim E, Lim YA, et al. Comparison of multiplex reverse transcription polymerase chain reaction and conventional cytogenetics as a diagnostic strategy for acute leukemia. Int J Lab Hematol. 2008. 30:513–518.24. Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007. 35:e40.25. Kim YS, Kim GS, Lee CH, Choi SI, Rhang DW, Cho HC. The cytogenetic study of acute and chronic leukemic patients in Korea. Korean J Clin Pathol. 1997. 17:898–911.26. Ha JS, Ryoo NH, Jeon DS, Kim JR, Kim YJ. The cytogenetic analysis of leukemic patients; a study of 515 cases. Korean J Hematol. 2003. 38:8–14.27. Johansson B, Mertens F, Mitelman F. Geographic heterogeneity of neoplasia-associated chromosome aberrations. Genes Chromosomes Cancer. 1991. 3:1–7.28. King RL, Naghashpour M, Watt CD, Morrissette JJ, Bagg A. A comparative analysis of molecular genetic and conventional cytogenetic detection of diagnostically important translocations in more than 400 cases of acute leukemia, highlighting the frequency of false-negative conventional cytogenetics. Am J Clin Pathol. 2011. 135:921–928.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Detection of Residual Leukemia with Reverse Transcription-polymerase Chain Reaction from Patients with AML1/ETO Positive Acute Myeloid Leukemia in Remission
- The Utility of the Multiplex Reverse Transcriptase-Polymerase Chain Reaction Assay in the Detection of Hematologic Malignancies
- Development of Multiplex Reverse Transcription Polymerase Chain Reaction for Detection and Typing of Parainfluenza Viruses
- Two Concurrent Chromosomal Aberrations Involving Three-way t(3;21;8)(p21;q22;q22) and Two-way t(2;11)(q31;p15) Translocations in a Case of de novo Acute Myeloid Leukemia
- Simultaneous Detection and Identification of Human Respiratory Syncytial Virus, Influenza Virus A ( H3N2 , H1N1 ) and B by One - tube Multiplex Reverse Transcription Polymerase Chain Reaction