J Korean Med Sci.
2004 Oct;19(5):682-687. 10.3346/jkms.2004.19.5.682.
Genetic Alterations in Intrahepatic Cholangiocarcinoma as revealed by Degenerate Oligonucleotide Primed PCR-Comparative Genomic Hybridization
- Affiliations
-
- 1Institute of Human Genetics, Department of Anatomy, Brain Korea 21 Biomedical Sciences, Korea University College of Medicine, Seoul, Korea. parksh@korea.ac.kr
- 2Department of Pathology and Brain Korea 21 Project for Medical Science, Yonsei University College of Medicine, Seoul, Korea.
- 3Department of Anatomy, Medical Research Center, College of Medicine, Ewha Womans University, Seoul, Korea.
- KMID: 1733509
- DOI: http://doi.org/10.3346/jkms.2004.19.5.682
Abstract
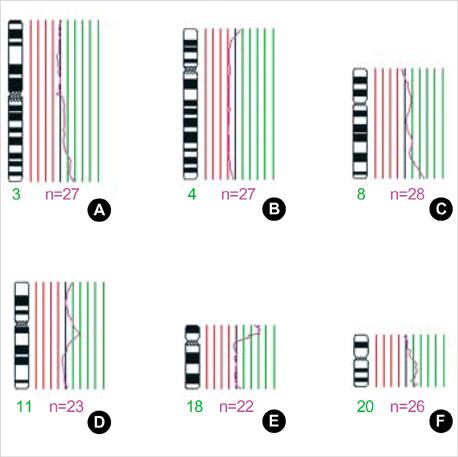
- Intrahepatic cholangiocarcinoma (ICC), a malignant neoplasm of the biliary epithelium, is usually fatal because of difficulty in early diagnosis and lack of availability of effective therapy. The genetic mechanisms involved in the development of ICC are not well understood and only a few cytogenetic studies of ICC have been published. Recently, technique of degenerate oligonucleotide primed (DOP)-PCR comparative genomic hybridization (CGH) permits genetic imbalances screening of the entire genome using only small amounts of tumor DNA. In this study chromosomal aberrations in 33 Korean ICC were investigated by DOP-PCR CGH. The common sites of copy number increases were 20q (67%), 17 (61%), 11q11-q13 (42%), 8p12-qter (39%), 18p (39%), 15q22-qter (36%), 16p (36%), 6p21 (30%), 3q25-qter (27%), 1q41-qter (24%), and 5p14-q11.2 (24%). DNA amplification was identified in 16 carcinomas (48%). The frequent sites of amplification were 20q, 17p, 17q23-qter, and 7p. The most frequent sites of copy number decreases were 1p32-pter (21%) and 4q (21%). The recurrent chromosomal aberrations identified in this study provide candidate regions involved in the tumorigenesis and progression of ICC.
Keyword
MeSH Terms
Figure
Reference
-
1. Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997. 13:405–409.
Article2. Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992. 13:718–725.
Article3. Huang Q, Schantz SP, Rao PH, Mo J, McCormick SA, Chaganti RS. Improving degenerate oligonucleotide primed PCR-comparative genomic hybridization for analysis of DNA copy number changes in tumors. Genes Chromosomes Cancer. 2000. 28:395–403.
Article4. Kim YI, Park CK, Kim JR, Chang JJ. Primary malignant epithelial neoplasms of the liver. J Korea Cancer Assoc. 1980. 12:33–53.5. Pederson LC, Buchsbaum DJ, Vickers SM, Kancharla SR, Mayo MS, Curiel DT, Stackhouse MA. Molecular chemotherapy combined with radiation therapy enhances killing of cholangiocarcinoma cells in vitro and in vivo. Cancer Res. 1997. 57:4325–4332.6. Holzinger F, Z'graggen K, Buchler MW. Mechanisms of biliary carcinogenesis: a pathogenetic multi-stage cascade towards cholangiocarcinoma. Ann Oncol. 1999. 10:Suppl 4. 122–126.
Article7. Koo SH, Ihm CH, Kwon KC, Park JW, Kim JM, Kong G. Genetic alterations in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Genet Cytogenet. 2001. 130:22–28.
Article8. Wong N, Li L, Tsang K, Lai PB, To KF, Johnson PJ. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol. 2002. 37:633–639.
Article9. Storto PD, Saidman SL, Demetris AJ, Letessier E, Whiteside TL, Gollin SM. Chromosomal breakpoint in cholangiocarcinoma cell line. Genes Chromosomes Cancer. 1990. 2:300–310.10. Kim DG, Park SY, You KR, Lee GB, Kim H, Moon WS, Chun YH, Park SH. Establishment and characterization of chromosomal aberrations in human cholangiocarcinoma cell lines by cross-species color banding. Genes Chromosomes Cancer. 2001. 30:48–56.
Article11. Kang YK, Kim YI, Kim WH. Allelotype analysis of intrahepatic cholangiocarcinoma. Mod Pathol. 2000. 13:627–631.
Article12. Kang YK, Kim WH, Lee HW, Lee HK, Kim YI. Mutation of p53 and K-ras, and loss of heterozygosity of APC in intrahepatic cholangiocarcinoma. Lab Invest. 1999. 79:477–483.13. Terada T, Ashida K, Endo K, Horie S, Maeta H, Matsunaga Y, Takashima K, Ohta T, Kitamura Y. C-erbB-2 protein is expressed in hepatolithiasis and cholangiocarcinoma. Histopathology. 1998. 33:325–331.
Article14. Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Kockerling F, Hauss J, Wittekind C. Frequency of p16INK4A alterations and k-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000. 47:721–727.15. Kuukasjarvi T, Tanner M, Pennanen S, Karhu R, Visakorpi T, Isola J. Optimizing DOP-PCR for universal amplification of small DNA samples in comparative genomic hybridization. Genes Chromosomes Cancer. 1997. 18:94–101.16. Kim GJ, Cho SJ, Won NH, Sung JM, Kim H, Chun YH, Park SH. Genomic imbalances in Korean hepatocellular carcinoma. Cancer Genet Cytogenet. 2003. 142:129–133.
Article17. el-Rifai W, Larramendy ML, Bjorkqvist AM, Hemmer S, Knuutila S. Optimazation of comparative genomic hybridization using fluorochrome conjugated to dCTP and dUTP nucleotides. Lab Invest. 1997. 77:699–700.18. Mahlamaki EH, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi OP, Johansson B. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p and 17q and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes Cancer. 1997. 20:383–391.19. Fukushige S, Waldman FM, Kimura M, Abe T, Furukawa T, Sunamura M, Kobari M, Horii A. Frequent gain of copy number on the long arm of chromosome 20 in human pancreatic adenocarcinoma. Genes Chromosomes Cancer. 1997. 19:161–169.
Article20. Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998. 21:177–184.
Article21. Saito M, Helin K, Valentine MB, Griffith BB, Willman CL, Harlow E, Look AT. Amplification of the E2F1 transcription factor gene in the HEL erythroleukemia cell line. Genomics. 1995. 25:130–138.
Article22. Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997. 277:965–968.
Article23. Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, Kuo WL, Yaswen P, Dairkee S, Giovanola J, Hutchinson GB, Isola J, Kallioniemi OP, Palazzolo M, Martin C, Ericsson C, Pinkel D, Gray JW, Albertson D, Li WB. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA. 1998. 95:8703–8708.
Article24. Ghadima BM, Schrock E, Walker RL, Wangsa D, Jauho A, Meltzer PS, Ried T. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am J Pathol. 1999. 154:525–536.25. Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998. 20:189–193.
Article26. Ukita Y, Kato M, Terada T. Gene amplification and mRNA and protein overexpression of c-erbB-2 (HER-2/neu) in human intrahepatic cholangiocarcinoma as detected by fluorescence in situ hybridization, in situ hybridization, and immunohistochemistry. J Hepatol. 2002. 36:780–785.
Article27. Meenakshi A, Kumar RS, Siva Kumar N, Genesh V. Immunofluorescent localization of C-erbB-2 oncoprotein in breast cancer: a preliminary study. Hum Antibodies. 2002. 11:73–77.
Article28. Takai S, Long JE, Yamada K, Miki T. Chromosomal localization of the human ECT2 proto-oncogene to 3q26.1→q26.2 by somatic cell analysis and fluorescence in situ hybridization. Genomics. 1995. 27:220–222.29. Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nature Genet. 1999. 21:99–102.
Article30. Byun DS, Cho K, Ryu BK, Lee MG, Park JI, Chae KS, Kim HJ, Chi SG. Frequent monoallelic deletion of PTEN and its reciprocal association with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003. 1004:318–327.31. Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001. 61:4122–4129.32. Feldman BJ, Reid TR, Cleary ML. Pim1 cooperates with E2a-Pbx1 to facilitate the progression of thymic lymphomas in transgenic mice. Oncogene. 1997. 15:2735–2742.
Article33. Inaba T, Matsushime H, Valentine M, Roussel MF, Sherr CJ, Look AT. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics. 1992. 13:565–574.
Article34. Akervall JA, Jin Y, Wennerberg JP, Zatterstrom UK, Kjellen E, Mertens F, Willen R, Mandahl N, Heim S, Mitelman F. Chromosomal abnormalities involving 11q13 are associated with poor prognosis in patients with squamous cell carcinoma of the head and neck. Cancer. 1995. 76:853–859.
Article35. Schwab M. Amplification of oncogenes in human cancer cells. Bioessays. 1998. 20:473–479.
Article36. Dickson C, Fantl V, Gillett C, Brookes S, Bartek J, Smith R, Fisher C, Barnes D, Peters G. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett. 1995. 90:43–50.
Article37. Schuuring E. The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes-a review. Gene. 1995. 159:83–96.
Article38. Piao Z, Park C, Park JH, Kim H. Allelotype analysis of hepatocellular carcinoma. Int J Cancer. 1998. 75:29–33.
Article39. Momoi H, Okabe H, Kamikawa T, Satoh S, Ikai I, Yamamoto M, Nakagawara A, Shimahara Y, Yamaoka Y, Fukumoto M. Comprehensive allelotyping of human intrahepatic cholangiocarcinoma. Clin Cancer Res. 2001. 7:2648–2655.40. Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, Yu XJ, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J Hepatol. 2001. 34:840–849.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Degenerate oligonucleotide primed PCR for the application to comparative genomic hybridization
- Genomic Imbalances in Ependymoma by Degenerate Oligonucleotide Primed PCR-Comparative Genomic Hybridization
- A study on the chromosomal aberrations in Korean intrahepatic cholangiocarcinomas with comparative genomic hybridization
- A New Prenatal Diagnosis of Fetal Cells Isolation from Maternal Peripheral Blood -Using Comparative Genomic Hybridization by Microdissection
- Determination of Chromosomal Alterations in Nasal NK/T-cell Lymphomas by DOP-PCR and Comparative Genomic Hybridization