Endocrinol Metab.
2011 Jun;26(2):177-184. 10.3803/EnM.2011.26.2.177.
Mutational Analysis of the NF1 Gene in Two Families with Neurofibromatosis 1 Accompanied by Pheochromocytoma
- Affiliations
-
- 1Department of Internal Medicine and Laboratory of Molecular Endocrinology, Gachon University School of Medicine, Incheon, Korea. shleemd@gachon.ac.kr
- 2Laboratory of Cancer Cell Biology, Lee Gil Ya Cancer and Diabetes Institute, Gachon University of Medicine and Science, Incheon, Korea.
- 3Department of Plastic Surgery, Gachon University School of Medicine, Incheon, Korea.
- KMID: 1497727
- DOI: http://doi.org/10.3803/EnM.2011.26.2.177
Abstract
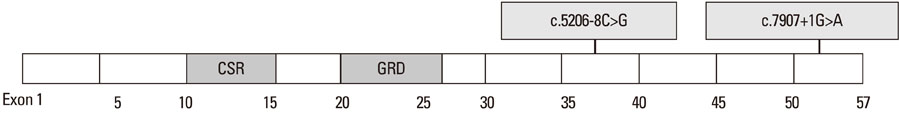
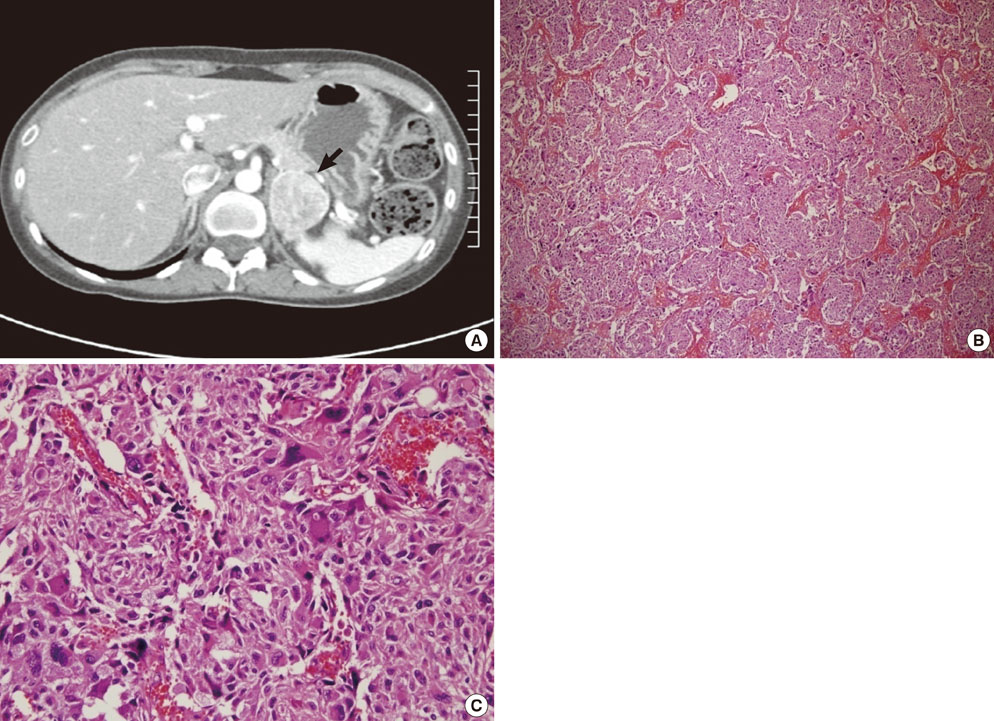
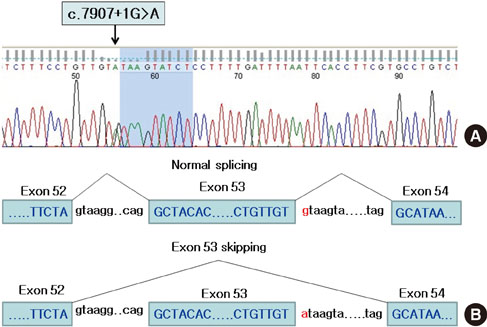
- Neurofibromatosis type 1 (NF1) is one of the most common autosomal dominant inherited disorders affecting the nervous system. NF1 is associated with mutations in the NF1 gene, which is located on chromosome sub-band 17q11.2 and contains 57 exons spanning approximately 300 kb of genomic DNA. NF1 is caused by a loss of function mutation of the NF1 gene, a tumor suppressor gene, which encodes for neurofibromin, a GTPase-activating protein (GAP) involved in the negative regulation of Ras activity. The GAP-related domain, which is encoded for by exons 20-27a, is one of the most important functional domains in neurofibromin. The cysteine-serine-rich domain has been recognized as an important functional domain in NF1-related pheochromocytomas. As the result of many genetic analyses of NF1-related pheochromocytomas, pheochromocytoma has generally been recognized as a true component of NF1. We recently experienced two families with NF1 accompanied by pheochromocytoma. The proband of family 1 is a 31-year-old female diagnosed with NF1 and pheochromocytoma. Gene analysis of the proband and her sister showed that the mutation of the NF1 gene (c.7907+1G>A) led to the skipping of exon 53 during NF1 mRNA splicing. The proband of family 2 is a 48-year-old male who was diagnosed with the same condition. Gene analysis demonstrated the mutation of the NF1 gene (c.5206-8C>G) with missplicing of exon 37. These novel germline mutations did not fall into the GAP-related nor the cysteine-serine-rich domains, but into the C-terminal area of the NF1 gene. This suggests that the correlation between the genotype and phenotype of NF1-related pheochromocytoma is somewhat difficult to characterize. Further studies will be necessary to confirm the function of the C-terminal area of the NF1 gene and its contribution to the development of NF1 and pheochromocytoma.
Keyword
MeSH Terms
-
Adult
DNA
Exons
Female
Genes, Neurofibromatosis 1
Genes, Tumor Suppressor
Genotype
Germ-Line Mutation
GTPase-Activating Proteins
Humans
Male
Middle Aged
Nervous System
Neurofibromatoses
Neurofibromatosis 1
Neurofibromin 1
Phenotype
Pheochromocytoma
RNA, Messenger
Siblings
DNA
GTPase-Activating Proteins
Neurofibromin 1
RNA, Messenger
Figure
Reference
-
1. Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, Robert W. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990. 62:599–608.2. Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubenstein A, Viskochil D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997. 278:51–57.3. Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, White R, O'Connell P. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990. 62:187–192.4. Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992. 356:713–715.5. DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992. 69:265–273.6. Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, Innis MA, McCormick F. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990. 63:843–849.7. Jeong SY, Park SJ, Kim HJ. The spectrum of NF1 mutations in Korean patients with neurofibromatosis type 1. J Korean Med Sci. 2006. 21:107–112.8. Lazaro C, Gaona A, Estivill X. Two CA/GT repeat polymorphisms in intron 27 of the human neurofibromatosis (NF1) gene. Hum Genet. 1994. 93:351–352.9. Xu GF, Nelson L, O'Connell P, White R. An Alu polymorphism intragenic to the neurofibromatosis type 1 gene (NF1). Nucleic Acids Res. 1991. 19:3764.10. Lazaro C, Gaona A, Xu G, Weiss R, Estivill X. A highly informative CA/GT repeat polymorphism in intron 38 of the human neurofibromatosis type 1 (NF1) gene. Hum Genet. 1993. 92:429–430.11. Lopez Correa C, Brems H, Lazaro C, Estivill X, Clementi M, Mason S, Rutkowski JL, Marynen P, Legius E. Molecular studies in 20 submicroscopic neurofibromatosis type 1 gene deletions. Hum Mutat. 1999. 14:387–393.12. Gutmann DH, Cole JL, Stone WJ, Ponder BA, Collins FS. Loss of neurofibromin in adrenal gland tumors from patients with neurofibromatosis type I. Genes Chromosomes Cancer. 1994. 10:55–58.13. Wimmer K, Roca X, Beiglbock H, Callens T, Etzler J, Rao AR, Krainer AR, Fonatsch C, Messiaen L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5' splice-site disruption. Hum Mutat. 2007. 28:599–612.14. Pros E, Gomez C, Martin T, Fabregas P, Serra E, Lazaro C. Nature and mRNA effect of 282 different NF1 point mutations: focus on splicing alterations. Hum Mutat. 2008. 29:E173–E193.15. Thomas SL, Deadwyler GD, Tang J, Stubbs EB Jr, Muir D, Hiatt KK, Clapp DW, De Vries GH. Reconstitution of the NF1 GAP-related domain in NF1-deficient human Schwann cells. Biochem Biophys Res Commun. 2006. 348:971–980.16. Bausch B, Borozdin W, Mautner VF, Hoffmann MM, Boehm D, Robledo M, Cascon A, Harenberg T, Schiavi F, Pawlu C, Peczkowska M, Letizia C, Calvieri S, Arnaldi G, Klingenberg-Noftz RD, Reisch N, Fassina A, Brunaud L, Walter MA, Mannelli M, MacGregor G, Palazzo FF, Barontini M, Walz MK, Kremens B, Brabant G, Pfaffle R, Koschker AC, Lohoefner F, Mohaupt M, Gimm O, Jarzab B, McWhinney SR, Opocher G, Januszewicz A, Kohlhase J, Eng C, Neumann HP. Germline NF1 mutational spectra and loss-of-heterozygosity analyses in patients with pheochromocytoma and neurofibromatosis type 1. J Clin Endocrinol Metab. 2007. 92:2784–2792.17. Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000. 403:895–898.18. Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002. 5:95–96.19. Dasgupta B, Dugan LL, Gutmann DH. The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J Neurosci. 2003. 23:8949–8954.20. Pros E, Fernandez-Rodriguez J, Canet B, Benito L, Sanchez A, Benavides A, Ramos FJ, Lopez-Ariztegui MA, Capella G, Blanco I, Serra E, Lazaro C. Antisense therapeutics for neurofibromatosis type 1 caused by deep intronic mutations. Hum Mutat. 2009. 30:454–462.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Spectrum of NF1 Mutations in Korean Patients with Neurofibromatosis Type 1
- A novel neurofibromatosis type 1 (NF1) mutation in a patient with NF1 and pheochromocytoma
- Mutation spectrum of NF1 gene in Korean unrelated patients with neurofibromatosis 1: Six novel pathogenic variants
- Mutation of the NF1 Gene and the Associated Clinical Features in Family Members with Neurofibromatosis Type 1
- Neurofibromatosis Type I: Case Reports of Two Novel Mutations