Lab Anim Res.
2010 Mar;26(1):91-94. 10.5625/lar.2010.26.1.91.
New Splicing Variants of the Murine Damaged DNA Binding 2
- Affiliations
-
- 1Department of Medical Genetics, College of Medicine, Hallym University, Chuncheon, Korea. jgsuh@hallym.ac.kr
- 2Genetics of Development and Disease Branch, NIH/NIDDK, USA.
- 3Institute of Natural Medicine, Hallym University, Chuncheon, Korea.
- 4Biosafty Research Institute, College of Medicine, Chonbuk National University, Jeonju, Korea.
- KMID: 1458945
- DOI: http://doi.org/10.5625/lar.2010.26.1.91
Abstract
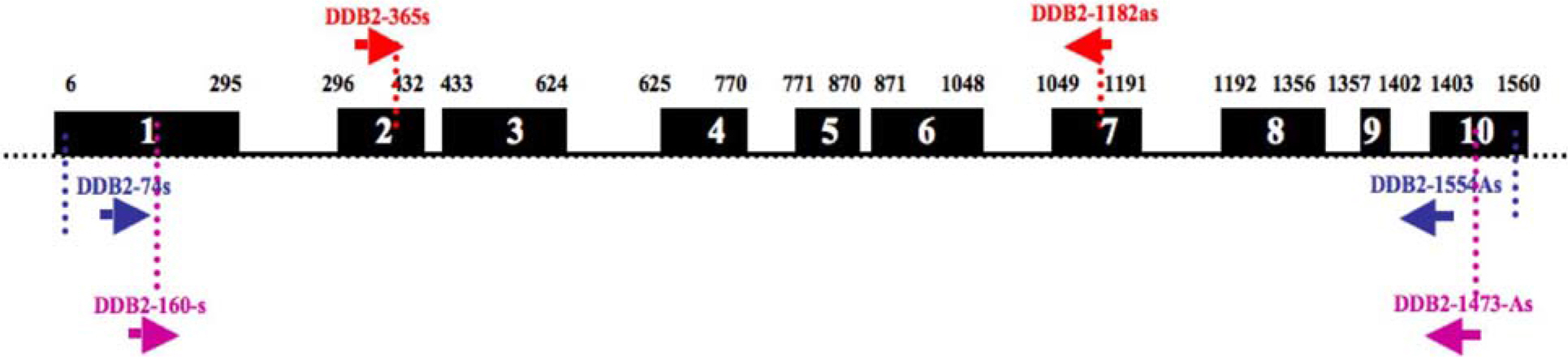
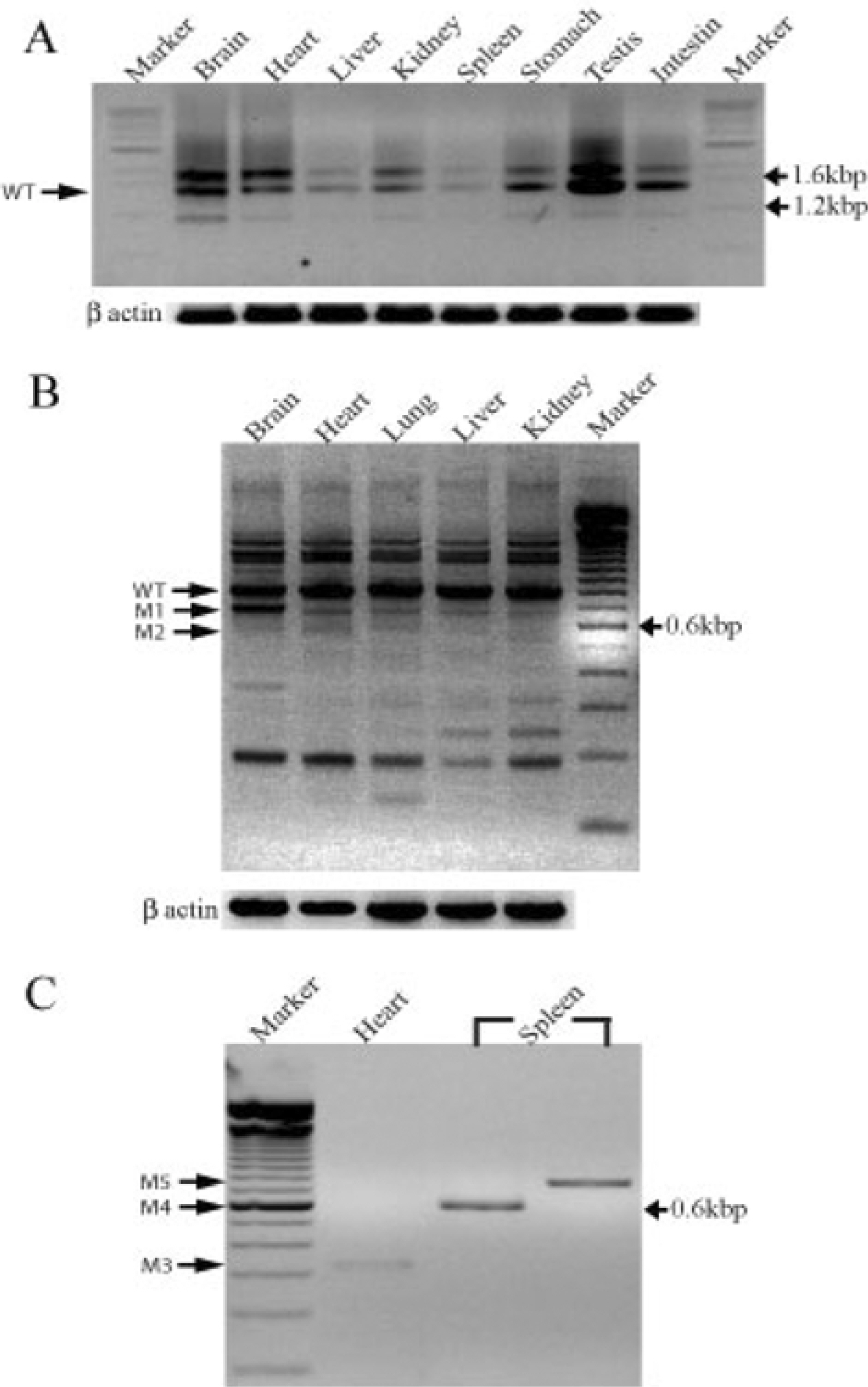
- Damaged DNA binding (DDB) protein is an important gene in the repair of damaged DNA. DDB is a heterodimer (DDB1 and DDB2) protein, murine DDB2 has 10 exons about 1.5kb in size (Genbank Accession No. AY027937). Here we identified five DDB2 variants (M1-M5) from various mouse tissues that are generated by alternative splicing. We used reverse transcription-PCR (RT-PCR) to identify splicing variants and isolated PCR products using an agarose-gel PCR purification kit. All isolated PCR products were cloned and the structure of splicing variants was confirmed by sequencing. The first splicing variant M1 was generated by omission of exon 4. The second splicing variant M2, by omission of exons 4-5. The third variants M3 was generated by omission from the middle of exon 1 to exon 6 and was expressed in the heart. Fourth variants M4 was generated by omission of exon 2 and exons 4-7. M5, the last splicing variant was generated by omission of exons 4-7. M4 and M5 were expressed in the spleen. Analysis of tissue distribution by RT-PCR indicates that M1 is most highly expressed in the mouse brain. These results indicated that murine DDB2 has five splicing variants and splicing variants expression patterns were different depending on mouse tissue. Further functional studies of each splicing variants will provide more information about the molecular mechanism of DDB2 function and DDB2 gene expression regulation.
Keyword
MeSH Terms
Figure
Reference
-
Aboussekhra A.., Biggerstaff M.., Shivji M.K.., Vilpo J.A.., Moncollin V.., Podust V.N.., Protic M.., Hubscher U.., Egly J.M.., Wood R.D.1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 80(6):859–868.
ArticleBlack D.L.2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291–336.
ArticleBohr V.A.., Smith C.A.., Okumoto D.S.., Hanawalt P.C.1985. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 40(2):359–369.
ArticleDavid C.J.., Manley J.L.2008. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 22(3):279–285.
ArticleFedor M.J.2008. Alternative splicing minireview series: combinatorial control facilitates splicing regulation of gene expression and enhances genome diversity. J. Biol. Chem. 283(3):1209–1210.
ArticleFujiwara Y.., Masutani C.., Mizukoshi T.., Kondo J.., Hanaoka F.., Iwai S.1999. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 274(28):20027–20033.
ArticleGraveley B.R.2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17(2):100–107.
ArticleGraveley B.R.2009. Alternative splicing: regulation without regulators. Nat. Struct. Mol. Biol. 16(1):13–15.
ArticleHwang B.J.., Ford J.M.., Hanawalt P.C.., Chu G.1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA. 96(2):424–428.
ArticleInoki T.., Yamagami S.., Inoki Y.., Tsuru T.., Hamamoto T.., Kagawa Y.., Mori T.., Endo H.2004. Human DDB2 splicing variants are dominant negative inhibitors of UV-damaged DNA repair. Biochem. Biophys. Res. Commun. 314(4):1036–1043.
ArticleItoh M.., Nagatomo K.., Kubo Y.., Saitoh O.2006. Alternative splicing of RGS8 gene changes the binding property to the M1 muscarinic receptor to confer receptor type-specific Gq regulation. J. Neurochem. 99(6):1505–1516.
ArticleItoh T.., Cado D.., Kamide R.., Linn S.2004. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc. Natl. Acad. Sci. USA. 101(7):2052–2057.
ArticleItoh T.., Mori T.., Ohkubo H.., Yamaizumi M.1999. A newly identified patient with clinical xeroderma pigmentosum phenotype has a non-sense mutation in the DDB2 gene and incomplete repair in (6-4) photoproducts. J. Invest. Dermatol. 113(2):251–257.
ArticleKeeney S.., Chang G.J.., Linn S.1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268(28):21293–21300.
ArticleMayeda A.., Helfman D.M.., Krainer A.R.1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell Biol. 13(5):2993–3001.
ArticleOhno G.., Hagiwara M.., Kuroyanagi H.2008. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 22(3):360–374.
ArticleRoesser J.R.., Liittschwager K.., Leff S.E.1993. Regulation of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide gene by RNA-binding proteins. J. Biol. Chem. 268(11):8366–8375.
ArticleStoyanova T.., Roy N.., Kopanja D.., Raychaudhuri P.., Bagchi S.2009. DDB2 (Damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response. Cell Cycle. 8(24):4067–4071.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Alternative Splicing and Its Impact as a Cancer Diagnostic Marker
- Application of a Repair Enzyme to STR Analysis of Damaged DNA
- The Pathogenetic Role of TAR DNA Binding Protein (TDP-43) in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia
- A Review of Deep Genomics Applying Machine Learning in Genomic Medicine
- Reclassification of Five BRCA1/2 Variants with Unknown Significance Using Complex Functional Study