J Korean Assoc Oral Maxillofac Surg.
2012 Feb;38(1):14-19. 10.5125/jkaoms.2012.38.1.14.
Expression of ssrA in non-pathogen-induced adaptation in the oral cavity through signal exchange with oral pathogens
- Affiliations
-
- 1Department of Oral and Maxillofacial Surgery, School of Dentistry, Dental Science Research Institute, 2nd Stage of Brain Korea 21, Chonnam National University, Gwangju, Korea. omspark@chonnam.ac.kr
- KMID: 1434374
- DOI: http://doi.org/10.5125/jkaoms.2012.38.1.14
Abstract
- INTRODUCTION
This study was conducted to evaluate ssrA expression resulting from adaptation of Escherichia coli (E. coli) to oral pathogens through signal exchange.
MATERIALS AND METHODS
Human cell lines Hep2 and HT29, wild-type E. coli (WT K-12), ssrA knock-out E. coli (Delta K-12), and Scleropages aureus (S. aureus) were used. A single culture consisting of Hep2, HT29, WT K-12, and Delta K-12, and mixed cultures consisting of Hep2 and WT K-12, Hep2 and Delta K-12, WT K-12 and S. aureus , Delta K-12 and S. aureus , and Hep2, WT K-12, and S. aureus were prepared. For HT29, a mixed culture was prepared with WT K-12 and with WT K-12 and S. aureus. Total RNA was extracted from each culture with the resulting expression of ssrA, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappaB), and p53 was evaluated by Reverse transcription polymerase chain reaction (RT-PCR).
RESULTS
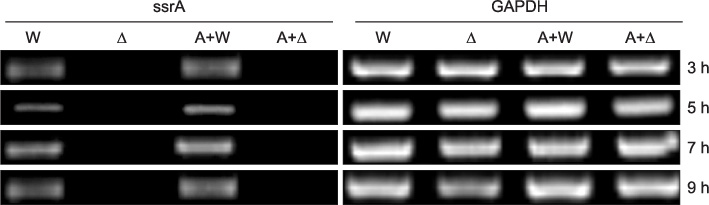
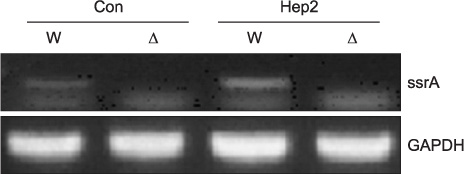
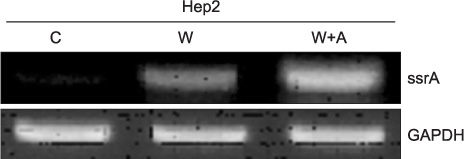
The expression of ssrA in a single culture of WT K-12 was lower than that observed in the mixed culture of WT K-12 with S. aureus. Greater ssrA expression was observed in the mixed culture of WT K-12 with Hep2 than in the single culture of WT K-12. The expression of NF-kappaB was higher in the mixed culture of Hep2 with Delta K-12 than that in the mixed culture of Hep2 with WT K-12, and was lowest in the single culture of Hep2. The expression of ssrA was higher in the mixed culture of WT K-12 with Hep2 and S. aureus than in the mixed culture of WT K-12 with Hep2.
CONCLUSION
These results suggest that ssrA plays an important role in the mechanism of E. coli adaptation to a new environment.
Keyword
MeSH Terms
Figure
Reference
-
1. Isselbacher KJ, Braunwald E, Wilson JD, Martin JB, Fauci AS, Kasper DL. Harrison's principles of internal medicine. 1997. 13th edition. New York: McGraw-Hill.2. Frisken KW, Higgins T, Palmer JM. The incidence of periodontopathic microorganisms in young children. Oral Microbiol Immunol. 1990. 5:43–45.
Article3. Slots J, Genco RJ. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984. 63:412–421.
Article4. Moore WE. Microbiology of periodontal disease. J Periodontal Res. 1987. 22:335–341.
Article5. Genco RJ, Zambon JJ, Christersson LA. The origin of periodontal infections. Adv Dent Res. 1988. 2:245–259.
Article6. Braiteh F, Golden MP. Cryptogenic invasive Klebsiella pneumoniae liver abscess syndrome. Int J Infect Dis. 2007. 11:16–22.
Article7. Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005. 38:135–187.
Article8. Kirby JE, Trempy JE, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994. 176:2068–2081.
Article9. Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996. 271:990–993.
Article10. Retallack DM, Johnson LL, Friedman DI. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J Bacteriol. 1994. 176:2082–2089.
Article11. Withey J, Friedman D. Analysis of the role of trans-translation in the requirement of tmRNA for lambdaimmP22 growth in Escherichia coli. J Bacteriol. 1999. 181:2148–2157.
Article12. Retallack DM, Friedman DI. A role for a small stable RNA in modulating the activity of DNA-binding proteins. Cell. 1995. 83:227–235.
Article13. Karzai AW, Roche ED, Sauer RT. The SsrA -SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000. 7:449–455.
Article14. Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol. 2003. 57:101–123.15. Fujihara A, Tomatsu H, Inagaki S, Tadaki T, Ushida C, Himeno H, et al. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells. 2002. 7:343–350.
Article16. Wiegert T, Schumann W. SsrA -mediated tagging in Bacillus subtilis. J Bacteriol. 2001. 183:3885–3889.17. Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1994. 91:9223–9227.
Article18. Muto A, Fujihara A, Ito KI, Matsuno J, Ushida C, Himeno H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells. 2000. 5:627–635.
Article19. Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994. 62:1623–1630.20. Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000. 182:1558–1563.
Article21. Kovach ME, Shaffer MD, Peterson KM. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996. 142:2165–2174.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Oral Pathogens and Their Antibiotics from Marine Organisms: A Systematic Review of New Drugs for Novel Drug Targets
- Role of Hepatocyte Growth Factor and c-met Gene Expression in Oral Cavity and Oropharyngeal Squamous Cell Carcinoma
- Oral cavity cancer management during the COVID-19 pandemic
- Comparison of Oral Hygiene Effects between 0.1% Chlorhexidine and Normal Saline on the Incidence of Oral Pathogens
- Surgical Excision and Reconstruction in Oral Cavity Cancer