Ann Lab Med.
2012 Jul;32(4):257-263. 10.3343/alm.2012.32.4.257.
Evaluation of Peptide Nucleic Acid Probe-based Real-time PCR for Detection of Mycobacterium tuberculosis Complex and Nontuberculous Mycobacteria in Respiratory Specimens
- Affiliations
-
- 1Department of Laboratory Medicine, Soonchunhyang University College of Medicine, Cheonan, Korea. clinpath@sch.ac.kr
- 2Department of Clinical Parasitology and Allergy, Soonchunhyang University College of Medicine, Cheonan, Korea.
- 3Department of Biochemistry, Soonchunhyang University College of Medicine, Cheonan, Korea.
- 4Department of Pediatrics, Soonchunhyang University College of Medicine, Cheonan, Korea.
- 5Chungcheongnam-Do Health and Environment Research Institute, Daejeon, Korea.
- KMID: 1380084
- DOI: http://doi.org/10.3343/alm.2012.32.4.257
Abstract
- BACKGROUND
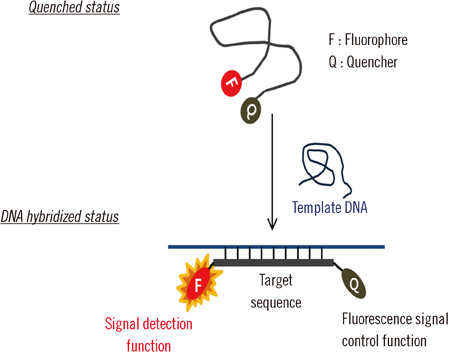
A peptide nucleic acid (PNA) probe-based real-time PCR (PNAqPCR(TM) TB/NTM detection kit; PANAGENE, Korea) assay has been recently developed for the simultaneous detection of Mycobacterium tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM) in clinical specimens. The study was aimed at evaluation of the performance of PNA probe-based real-time PCR in respiratory specimens.
METHODS
To evaluate potential cross-reactivity, the extracted DNA specimens from Mycobacterium species and non-mycobacterial species were tested using PNA probe-based real-time PCR assay. A total of 531 respiratory specimens (482 sputum specimens and 49 bronchoalveolar washing fluid specimens) were collected from 230 patients in July and August, 2011. All specimens were analyzed for the detection of mycobacteria by direct smear examination, mycobacterial culture, and PNA probe-based real-time PCR assay.
RESULTS
In cross-reactivity tests, no false-positive or false-negative results were evident. When the culture method was used as the gold standard test for comparison, PNA probe-based real-time PCR assay for detection of MTBC had a sensitivity and specificity of 96.7% (58/60) and 99.6% (469/471), respectively. Assuming the combination of culture and clinical diagnosis as the standard, the sensitivity and specificity of the new real-time PCR assay for detection of MTBC were 90.6% (58/64) and 99.6% (465/467), respectively. The new real-time PCR for the detection of NTM had a sensitivity and specificity of 69.0% (29/42) and 100% (489/489), respectively.
CONCLUSIONS
The new real-time PCR assay may be useful for the detection of MTBC in respiratory specimens and for discrimination of NTM from MTBC.
MeSH Terms
-
Bronchoalveolar Lavage Fluid/microbiology
DNA Probes/chemistry/metabolism
DNA, Bacterial/*analysis
Humans
Molecular Typing/*methods
Mycobacterium tuberculosis/*genetics/isolation & purification
Nontuberculous Mycobacteria/*genetics/isolation & purification
Nucleic Acid Hybridization
Peptide Nucleic Acids/chemistry/*metabolism
*Real-Time Polymerase Chain Reaction
Respiratory System/*microbiology
Sputum/microbiology
Figure
Reference
-
1. WHO Report 2010 global tuberculosis control. World Health Organization. Updated on Sep 2011. http://whqlibdoc.who.int/publications/2010/9789241564069_eng.pdf.2. Peter-Getzlaff S, Lüthy J, Böddinghaus B, Böttger EC, Springer B. Development and evaluation of a molecular assay for detection of nontuberculous mycobacteria by use of the cobas amplicor platform. J Clin Microbiol. 2008. 46:4023–4028.
Article3. Primm TP, Lucero CA, Falkinham JO 3rd. Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004. 17:98–106.
Article4. Richter E, Brown-Elliot BA, Wallace RJ. Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Mycobacterium: laboratory characteristics of slowly growing mycobacteria. Manual of clinical microbiology. 2011. 10th ed. Washington, DC: ASM Press;503–524.5. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007. 175:367–416.
Article6. Shinnick TM, Iademarco MF, Ridderhof JC. National plan for reliable tuberculosis laboratory services using a systems approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR Recomm Rep. 2005. 54:1–12.7. Forbes EA. Persing DH, Tenover FC, Tang YW, Nolte FS, Hayden RT, van Belkum A, editors. Molecular detection and characterization of Mycobacterium tuberculosis. Molecular microbiology: diagnostic principles and practice. 2011. 2nd ed. Washington, DC: ASM Press;415–436.8. Apers L, Mutsvangwa J, Magwenzi J, Chigara N, Butterworth A, Mason P, et al. A comparison of direct microscopy, the concentration method and the Mycobacteria Growth Indicator Tube for the examination of sputum for acid-fast bacilli. Int J Tuberc Lung Dis. 2003. 7:376–381.9. Fitzgerald DW, Sterling TR, Haas DW. Mandell GL, Bennett JE, Dolin R, editors. Mycobacterium tuberculosis. Mandell, Douglas, and Bennett's Principles and practice of infectious disease. 2010. 7th ed. Philadelphia: Churchill Livingstone Elsevier;3129–3163.
Article10. Centers for Disease Control and Prevention. Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009. 58:7–10.11. Drouillon V, Delogu G, Dettori G, Lagrange PH, Benecchi M, Houriez F, et al. Multicenter evaluation of a transcription-reverse transcription concerted assay for rapid detection of Mycobacterium tuberculosis complex in clinical specimens. J Clin Microbiol. 2009. 47:3461–3465.12. Kim K, Lee H, Lee MK, Lee SA, Shim TS, Lim SY, et al. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J Clin Microbiol. 2010. 48:3073–3080.13. Porcheddu A, Giacomelli G. Peptide nucleic acids (PNAs), a chemical overview. Curr Med Chem. 2005. 12:2561–2599.
Article14. Choi YJ, Kim HS, Lee SH, Park JS, Nam HS, Kim HJ, et al. Evaluation of peptide nucleic acid array for the detection of hepatitis B virus mutations associated with antiviral resistance. Arch Virol. 2011. 156:1517–1524.
Article15. Park H, Jang H, Song E, Chang CL, Lee M, Jeong S, et al. Detection and genotyping of Mycobacterium species from clinical isolates and specimens by oligonucleotide array. J Clin Microbiol. 2005. 43:1782–1788.16. Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008. 14:2439–2444.
Article17. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. Evaluation of the performances of AdvanSure TB/NTM real time PCR kit for detection of mycobacteria in respiratory specimens. Korean J Lab Med. 2008. 28:34–38.
Article18. Behr MA, Warren SA, Salamon H, Hopewell PC, Ponce de Leon A, Daley CL, et al. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet. 1999. 353:444–449.19. Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, et al. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess. 2007. 11:1–196.
Article20. Greco S, Girardi E, Navarra A, Saltini C. Current evidence on diagnostic accuracy of commercially based nucleic acid amplification tests for the diagnosis of pulmonary tuberculosis. Thorax. 2006. 61:783–790.
Article21. Marras TK, Chedore P, Ying AM, Jamieson F. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997-2003. Thorax. 2007. 62:661–666.22. Miguez-Burbano MJ, Flores M, Ashkin D, Rodriguez A, Granada AM, Quintero N, et al. Non-tuberculous mycobacteria disease as a cause of hospitalization in HIV-infected subjects. Int J Infect Dis. 2006. 10:47–55.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of Peptide Nucleic Acid Probe-Based Fluorescence In Situ Hybridization for the Detection of Mycobacterium tuberculosis Complex and Nontuberculous Mycobacteria in Clinical Respiratory Specimens
- Evaluation of the Diagnostic Performance of the AdvanSure TB/NTM Real-Time PCR Kit for Detection of Mycobacteria
- Recent Advances in Tuberculosis and Nontuberculous Mycobacteria Lung Disease
- Clinical Usefulness of Real-time PCR and Amplicor MTB PCR Assays for Diagnosis of Tuberculosis
- Evaluation of the AdvanSure TB/NTM Plus Real-Time PCR Assay for the Simultaneous Detection of Mycobacterium tuberculosis and Nontuberculous Mycobacteria from Clinical Specimens