J Korean Med Sci.
2025 May;40(19):e68. 10.3346/jkms.2025.40.e68.
Estimating the Prevalence of Autosomal Recessive Neuromuscular Diseases in the Korean Population
- Affiliations
-
- 1Department of Neurology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 2Department of Neurology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 3Rehabilitation Institute of Neuromuscular Disease, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2568104
- DOI: http://doi.org/10.3346/jkms.2025.40.e68
Abstract
- Background
Genetic neuromuscular diseases (NMDs) are a heterogeneous group of conditions that primarily affect the peripheral nerves, muscles, and neuromuscular junctions. This study was performed to identify pathogenic or likely pathogenic variants (PLPVs), calculate carrier frequencies, and predict the genetic prevalence of autosomal recessive-NMDs (AR-NMDs) in a Korean population.
Methods
In total, 267 genes were associated with AR-NMDs. We analyzed genetic variants from 984 Korean whole genomes and identified PLPVs to assess the carrier frequency and genetic prevalence of the variants.
Results
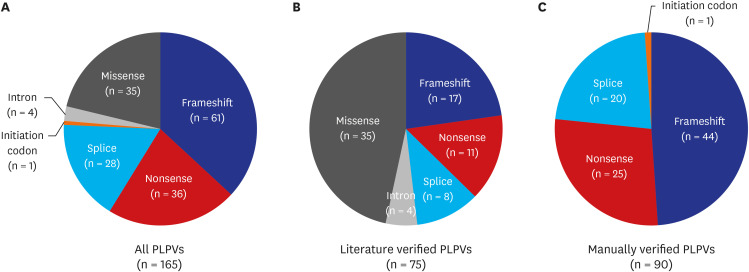
We identified 165 PLPVs, including 75 literature verified and 90 manually verified variants. Most PLPVs in AR-NMD genes were frameshifts (61, 37.0%), followed by nonsense (36, 21.8%), missense (35, 21.2%), and splice variants (28, 17.0%). The carrier frequency of the AR-NMDs was 27.1%. DYSF exhibited the highest carrier frequency (1.63%), followed by GAA (1.55%), HEXB (1.53%), PREPL (0.76%), NEB (0.66%), ADSS1 (0.65%), ALPK3 (0.65%), and CHRNG (0.65%). The predicted genetic prevalence of AR-NMDs in the Korean population was 38.0 cases per 100,000 individuals. DYSF (6.7 cases per 100,000 individuals) showed the highest genetic prevalence. The variant with the highest allele frequency was c.1250C>T in HEXB at 0.00764, followed by c.[752T>C; c.761C>T] in GAA at 0.00505, and c.2055+2T>G in DYSF at 0.00437.
Conclusion
Our study suggests that 27.1% of the Korean population are healthy carriers of at least one AR-NMD causing PLPV, revealing the genetic prevalence of NMDs in the Korean population.
Keyword
Figure
Reference
-
1. Benarroch L, Bonne G, Rivier F, Hamroun D. The 2024 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscul Disord. 2024; 34:126–170. PMID: 38253411.2. Savitz DA, Poole C, Miller WC. Reassessing the role of epidemiology in public health. Am J Public Health. 1999; 89(8):1158–1161. PMID: 10432898.3. Zhang L, Jin Y, Li J, He Z, Zhang D, Zhang M, et al. Epidemiological research on rare diseases using large-scale online search queries and reported case data. Orphanet J Rare Dis. 2023; 18(1):236. PMID: 37559136.4. Bhatt JM. The epidemiology of neuromuscular diseases. Neurol Clin. 2016; 34(4):999–1021. PMID: 27720006.5. Punga AR, Maddison P, Heckmann JM, Guptill JT, Evoli A. Epidemiology, diagnostics, and biomarkers of autoimmune neuromuscular junction disorders. Lancet Neurol. 2022; 21(2):176–188. PMID: 35065040.6. Müller KI, Ghelue MV, Lund I, Jonsrud C, Arntzen KA. The prevalence of hereditary neuromuscular disorders in Northern Norway. Brain Behav. 2021; 11(1):e01948. PMID: 33185984.7. Theadom A, Rodrigues M, Roxburgh R, Balalla S, Higgins C, Bhattacharjee R, et al. Prevalence of muscular dystrophies: a systematic literature review. Neuroepidemiology. 2014; 43(3-4):259–268. PMID: 25532075.8. Hong JM, Choi YC, Shin S, Lee JH, Shin HY, Kim SM, et al. Prevalence and socioeconomic status of patients with genetic myopathy in Korea: a nationwide, population-based study. Neuroepidemiology. 2019; 53(1-2):115–120. PMID: 31203286.9. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020; 581:434–443. PMID: 32461654.10. Zhao T, Fan S, Sun L. The global carrier frequency and genetic prevalence of Upshaw-Schulman syndrome. BMC Genom Data. 2021; 22(1):50. PMID: 34789164.11. Hanany M, Rivolta C, Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc Natl Acad Sci U S A. 2020; 117(5):2710–2716. PMID: 31964843.12. Choi WJ, Kim SH, Lee SR, Oh SH, Kim SW, Shin HY, et al. Global carrier frequency and predicted genetic prevalence of patients with pathogenic sequence variants in autosomal recessive genetic neuromuscular diseases. Sci Rep. 2024; 14(1):3806. PMID: 38361118.13. Jeon S, Bhak Y, Choi Y, Jeon Y, Kim S, Jang J, et al. Korean genome project: 1094 Korean personal genomes with clinical information. Sci Adv. 2020; 6(22):eaaz7835. PMID: 32766443.14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17(5):405–424. PMID: 25741868.15. Park HJ, Jang H, Kim JH, Lee JH, Shin HY, Kim SM, et al. Discovery of pathogenic variants in a large Korean cohort of inherited muscular disorders. Clin Genet. 2017; 91(3):403–410. PMID: 27363342.16. Park HJ, Hong YB, Hong JM, Yun U, Kim SW, Shin HY, et al. Null variants in DYSF result in earlier symptom onset. Clin Genet. 2021; 99(3):396–406. PMID: 33215690.17. Park HJ, Hong JM, Suh GI, Shin HY, Kim SM, Sunwoo IN, et al. Heterogeneous characteristics of Korean patients with dysferlinopathy. J Korean Med Sci. 2012; 27(4):423–429. PMID: 22468107.18. Park KS. Two approaches for a genetic analysis of pompe disease: a literature review of patients with pompe disease and analysis based on genomic data from the general population. Children (Basel). 2021; 8(7):601. PMID: 34356580.19. Taverna S, Cammarata G, Colomba P, Sciarrino S, Zizzo C, Francofonte D, et al. Pompe disease: pathogenesis, molecular genetics and diagnosis. Aging (Albany NY). 2020; 12(15):15856–15874. PMID: 32745073.20. Park HJ, Hong YB, Choi YC, Lee J, Kim EJ, Lee JS, et al. ADSSL1 mutation relevant to autosomal recessive adolescent onset distal myopathy. Ann Neurol. 2016; 79(2):231–243. PMID: 26506222.21. Saito Y, Nishikawa A, Iida A, Mori-Yoshimura M, Oya Y, Ishiyama A, et al. ADSSL1 myopathy is the most common nemaline myopathy in Japan with variable clinical features. Neurology. 2020; 95(11):e1500–e1511. PMID: 32646962.22. Soontrapa P, Liewluck T. Anoctamin 5 (ANO5) muscle disorders: a narrative review. Genes (Basel). 2022; 13(10):1736. PMID: 36292621.23. Cai S, Gao M, Xi J, Liu Z, Yue D, Wu H, et al. Clinical spectrum and gene mutations in a Chinese cohort with anoctaminopathy. Neuromuscul Disord. 2019; 29(8):628–633. PMID: 31350120.24. Kida H, Sano K, Yorita A, Miura S, Ayabe M, Hayashi Y, et al. First Japanese case of muscular dystrophy caused by a mutation in the anoctamin 5 gene. Neurol Clin Neurosci. 2015; 3(4):150–152.25. Kadoya M, Ogata K, Suzuki M, Honma Y, Momma K, Yatabe K, et al. A Japanese male with a novel ANO5 mutation with minimal muscle weakness and muscle pain till his late fifties. Neuromuscul Disord. 2017; 27(5):477–480. PMID: 28214267.26. Bohlega S, Monies DM, Abulaban AA, Murad HN, Alhindi HN, Meyer BF. Clinical and genetic features of anoctaminopathy in Saudi Arabia. Neurosciences. 2015; 20(2):173–177. PMID: 25864073.27. Lahoria R, Winder TL, Lui J, Al-Owain MA, Milone M. Novel ANO5 homozygous microdeletion causing myalgia and unprovoked rhabdomyolysis in an Arabic man. Muscle Nerve. 2014; 50(4):610–613. PMID: 24889862.28. Abouelhoda M, Sobahy T, El-Kalioby M, Patel N, Shamseldin H, Monies D, et al. Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet Med. 2016; 18(12):1244–1249. PMID: 27124789.29. El Mouzan MI, Al Salloum AA, Al Herbish AS, Qurachi MM, Al Omar AA. Consanguinity and major genetic disorders in Saudi children: a community-based cross-sectional study. Ann Saudi Med. 2008; 28(3):169–173. PMID: 18500181.30. Martin HC, Jones WD, McIntyre R, Sanchez-Andrade G, Sanderson M, Stephenson JD, et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018; 362(6419):1161–1164. PMID: 30409806.31. Carey IM, Banchoff E, Nirmalananthan N, Harris T, DeWilde S, Chaudhry UAR, et al. Prevalence and incidence of neuromuscular conditions in the UK between 2000 and 2019: a retrospective study using primary care data. PLoS One. 2021; 16(12):e0261983. PMID: 34972157.32. Emery AE. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul Disord. 1991; 1(1):19–29. PMID: 1822774.33. Cobben JM, van der Steege G, Grootscholten P, de Visser M, Scheffer H, Buys CH. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet. 1995; 57(4):805–808. PMID: 7573039.34. Chung CCY, Hue SPY, Ng NYT, Doong PHL, Chu ATW, et al. Hong Kong Genome Project. Meta-analysis of the diagnostic and clinical utility of exome and genome sequencing in pediatric and adult patients with rare diseases across diverse populations. Genet Med. 2023; 25(9):100896. PMID: 37191093.35. Fallin MD, Duggal P, Beaty TH. Genetic epidemiology and public health: the evolution from theory to technology. Am J Epidemiol. 2016; 183(5):387–393. PMID: 26905340.36. Westhoff CL. Epidemiologic studies: pitfalls in interpretation. Dialogues Contracept. 1995; 4(5):5–6. 8PMID: 12288680.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of Inheritance in Infantile Nystagmus
- Autosomal Recessive Polycystic Kidney Disease Confirmed to PKHD1 Gene Mutation: A Case of PKHD1 Gene Mutation
- Pulmonary hypertension in a child with juvenile-type autosomal recessive polycystic kidney disease
- Caroli Syndrome with Autosomal Recessive Polycystic Kidney Disease
- Homozygous Exon 4 Deletion in Parkin Gene in a Korean Family with Autosomal Recessive Early Onset Parkinsonism