Cancer Res Treat.
2025 Apr;57(2):492-506. 10.4143/crt.2024.667.
Presence of RB1 or Absence of LRP1B Mutation Predicts Poor Overall Survival in Patients with Gastric Neuroendocrine Carcinoma and Mixed Adenoneuroendocrine Carcinoma
- Affiliations
-
- 1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2566866
- DOI: http://doi.org/10.4143/crt.2024.667
Abstract
- Purpose
Neuroendocrine carcinomas (NECs) of the stomach are extremely rare, but fatal. However, our understanding of the genetic alterations in gastric NECs is limited. We aimed to evaluate genomic and clinicopathological characteristics of gastric NECs and mixed adenoneuroendocrine carcinomas (MANECs).
Materials and Methods
Fourteen gastric NECs, three gastric MANECs, and 1,381 gastric adenocarcinomas were retrieved from the departmental next-generation sequencing database between 2017 and 2022. Clinicopathological parameters and next-generation sequencing test results were retrospectively collected and reviewed.
Results
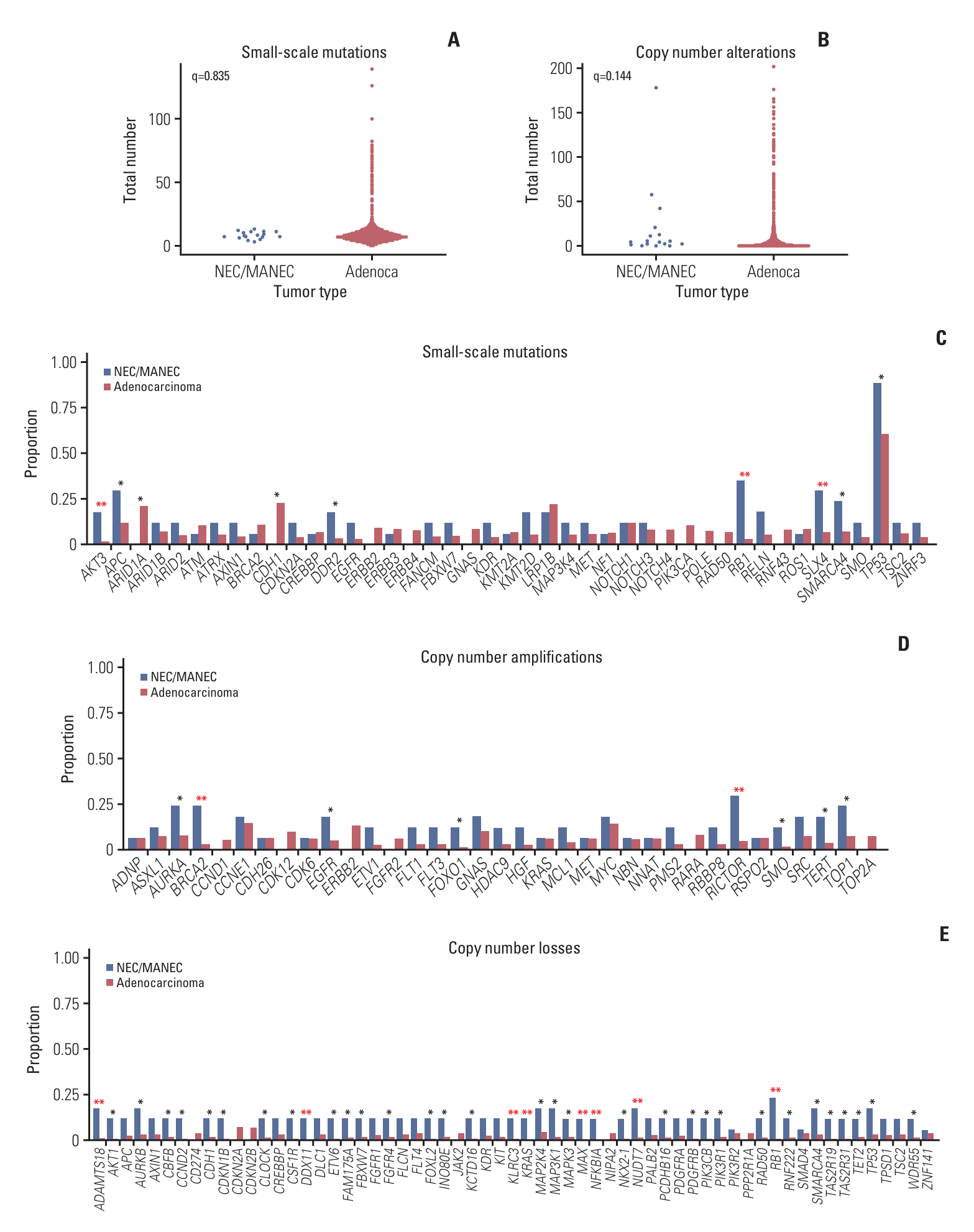
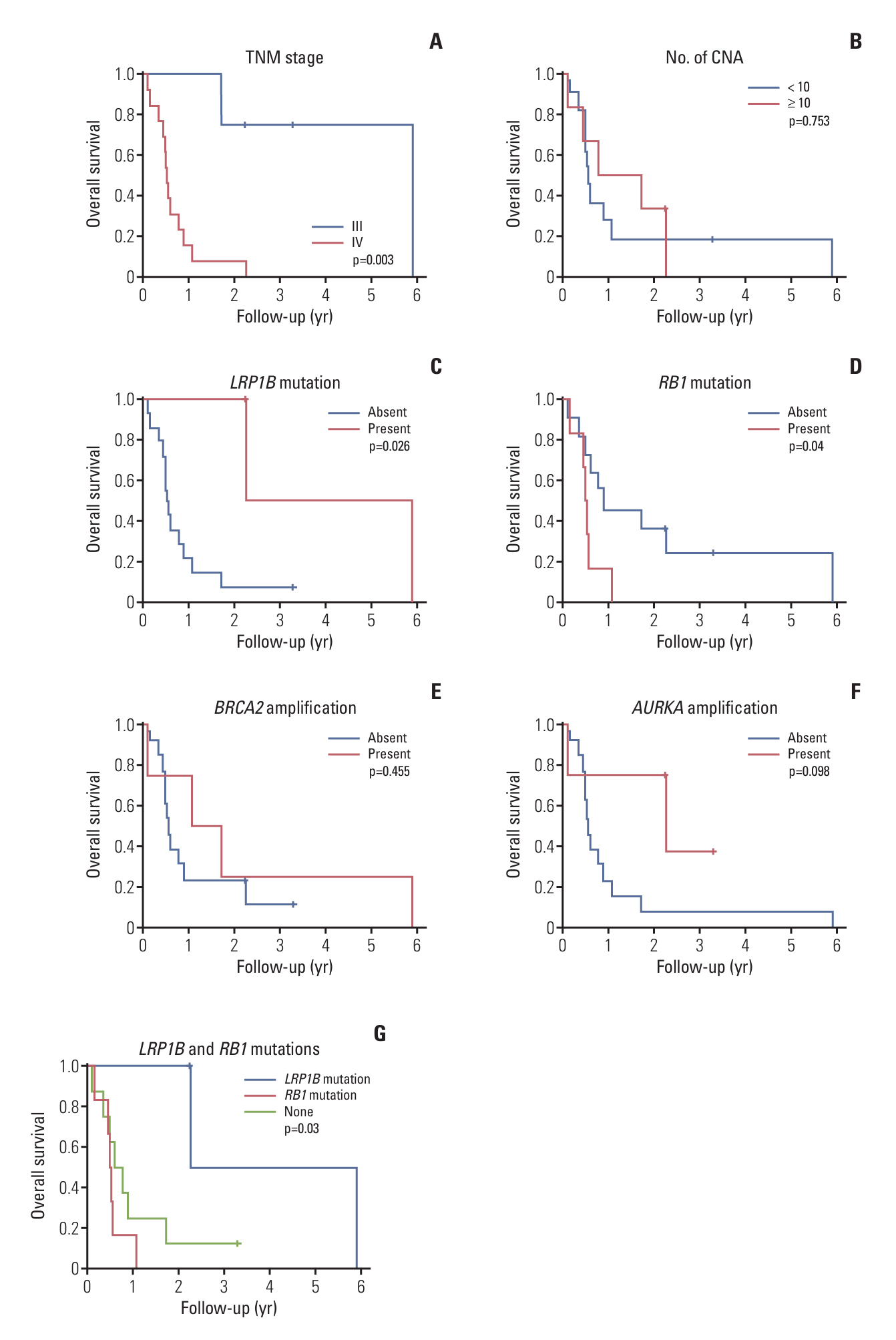
Gastric NECs and MANECs frequently harbored alterations of TP53, RB1, SMARCA4, RICTOR, APC, TOP1, SLX4, EGFR, BRCA2, and TERT. In contrast, gastric adenocarcinomas exhibited alterations of TP53, CDH1, LRP1B, ARID1A, ERBB2, GNAS, CCNE1, NOTCH, and MYC. Mutations of AKT3, RB1, and SLX4; amplification of BRCA2 and RICTOR; and deletion of ADAMTS18, DDX11, KLRC3, KRAS, MAX, NFKBIA, NUDT7, and RB1 were significantly more frequent in gastric NECs and MANECs than in gastric adenocarcinomas. The presence of LRP1B mutation was significantly associated with longer overall survival (OS), whereas RB1 mutation and advanced TNM stage were associated with shorter OS.
Conclusion
We identified frequently mutated genes and potential predictors of survival in patients with gastric NECs and MANECs.
Keyword
Figure
Reference
-
References
1. Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020; 76:182–8.
Article2. WHO Classification of Tumours Editorial Board. WHO classification of tumours: digestive system tumours. 5th ed. IARC Press;2019.3. Park JY, Ryu MH, Park YS, Park HJ, Ryoo BY, Kim MG, et al. Prognostic significance of neuroendocrine components in gastric carcinomas. Eur J Cancer. 2014; 50:2802–9.
Article4. Venizelos A, Elvebakken H, Perren A, Nikolaienko O, Deng W, Lothe IM, et al. The molecular characteristics of high-grade gastroenteropancreatic neuroendocrine neoplasms. Endocr Relat Cancer. 2021; 29:1–14.
Article5. Wu H, Yu Z, Liu Y, Guo L, Teng L, Guo L, et al. Genomic characterization reveals distinct mutation landscapes and therapeutic implications in neuroendocrine carcinomas of the gastrointestinal tract. Cancer Commun (Lond). 2022; 42:1367–86.
Article6. Koh J, Park HY, Bae JM, Kang J, Cho U, Lee SE, et al. Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists. J Pathol Transl Med. 2023; 57:265–72.
Article7. Kim JE, Chun SM, Hong YS, Kim KP, Kim SY, Kim J, et al. Mutation burden and I index for detection of microsatellite instability in colorectal cancer by targeted next-generation sequencing. J Mol Diagn. 2019; 21:241–50.
Article8. Oh JH, Sung CO, Kim HD, Chun SM, Kim J. BRCA-mutated gastric adenocarcinomas are associated with chromosomal instability and responsiveness to platinum-based chemotherapy. J Pathol Transl Med. 2023; 57:323–31.
Article9. Kim M, Lee C, Hong J, Kim J, Jeong JY, Park NJ, et al. Validation and clinical application of ONCOaccuPanel for targeted next-generation sequencing of solid tumors. Cancer Res Treat. 2023; 55:429–41.
Article10. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016; 44:e108.
Article11. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016; 12:e1004873.
Article12. Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015; 43:e19.
Article13. Robinson JT, Thorvaldsdottir H, Turner D, Mesirov JP. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics. 2023; 39:btac830.
Article14. Buchhalter I, Rempel E, Endris V, Allgauer M, Neumann O, Volckmar AL, et al. Size matters: Dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer. 2019; 144:848–58.
Article15. McClelland RA, Finlay P, Walker KJ, Nicholson D, Robertson JF, Blamey RW, et al. Automated quantitation of immunocytochemically localized estrogen receptors in human breast cancer. Cancer Res. 1990; 50:3545–50.16. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–9.
Article17. Guo X, Zhang B, Zeng W, Zhao S, Ge D. G3viz: an R package to interactively visualize genetic mutation data using a lollipop-diagram. Bioinformatics. 2020; 36:928–9.
Article18. Makuuchi R, Terashima M, Kusuhara M, Nakajima T, Serizawa M, Hatakeyama K, et al. Comprehensive analysis of gene mutation and expression profiles in neuroendocrine carcinomas of the stomach. Biomed Res. 2017; 38:19–27.
Article19. Wang H, Sun L, Bao H, Wang A, Zhang P, Wu X, et al. Genomic dissection of gastrointestinal and lung neuroendocrine neoplasm. Chin J Cancer Res. 2019; 31:918–29.
Article20. Koh J, Nam SK, Kwak Y, Kim G, Kim KK, Lee BC, et al. Comprehensive genetic features of gastric mixed adenoneuroendocrine carcinomas and pure neuroendocrine carcinomas. J Pathol. 2021; 253:94–105.
Article21. Ishida S, Akita M, Fujikura K, Komatsu M, Sawada R, Matsumoto H, et al. Neuroendocrine carcinoma and mixed neuroendocrine‒non-neuroendocrine neoplasm of the stomach: a clinicopathological and exome sequencing study. Hum Pathol. 2021; 110:1–10.
Article22. Yachida S, Totoki Y, Noe M, Nakatani Y, Horie M, Kawasaki K, et al. Comprehensive genomic profiling of neuroendocrine carcinomas of the gastrointestinal system. Cancer Discov. 2022; 12:692–711.
Article23. Griger J, Widholz SA, Jesinghaus M, de Andrade Kratzig N, Lange S, Engleitner T, et al. An integrated cellular and molecular model of gastric neuroendocrine cancer evolution highlights therapeutic targets. Cancer Cell. 2023; 41:1327–44.
Article24. Chen S, Sun L, Chen H, Li J, Lu C, Yang Y, et al. Clinicopathological and genetic characteristics of gastric neuroendocrine tumour (NET) G3 and comparisons with neuroendocrine carcinoma and NET G2. Histopathology. 2023; 83:700–11.
Article25. Qiu MZ, Chen Q, Zheng DY, Zhao Q, Wu QN, Zhou ZW, et al. Precise microdissection of gastric mixed adeno-neuroendocrine carcinoma dissects its genomic landscape and evolutionary clonal origins. Cell Rep. 2023; 42:112576.
Article26. Dieckmann M, Dietrich MF, Herz J. Lipoprotein receptors: an evolutionarily ancient multifunctional receptor family. Biol Chem. 2010; 391:1341–63.27. Principe C, Dionisio de Sousa IJ, Prazeres H, Soares P, Lima RT. LRP1B: a giant lost in cancer translation. Pharmaceuticals (Basel). 2021; 14:836.
Article28. He Z, Feng W, Wang Y, Shi L, Gong Y, Shi Y, et al. LRP1B mutation is associated with tumor immune microenvironment and progression-free survival in lung adenocarcinoma treated with immune checkpoint inhibitors. Transl Lung Cancer Res. 2023; 12:510–29.
Article29. Yuan T, Wang X, Sun S, Cao Z, Feng X, Gao Y. Profiling of 520 candidate genes in 50 surgically treated Chinese small cell lung cancer patients. Front Oncol. 2021; 11:644434.
Article30. Li J, Wei Q, Wu X, Sima J, Xu Q, Wu M, et al. Integrative clinical and molecular analysis of advanced biliary tract cancers on immune checkpoint blockade reveals potential markers of response. Clin Transl Med. 2020; 10:e118.
Article31. Innocenti F, Mu W, Qu X, Ou FS, Kabbarah O, Blanke CD, et al. DNA mutational profiling in patients with colorectal cancer treated with standard of care reveals differences in outcome and racial distribution of mutations. J Clin Oncol. 2024; 42:399–409.
Article32. Wang R, Zhang G, Zhu X, Xu Y, Cao N, Li Z, et al. Prognostic implications of LRP1B and its relationship with the tumor-infiltrating immune cells in gastric cancer. Cancers (Basel). 2023; 15:5759.
Article33. Xu J, Shen X, Zhang B, Su R, Cui M, Yan L, et al. Development and validation of LRP1B mutation-associated prognostic model for hepatocellular carcinoma. Biosci Rep. 2021; 41:BSR20211053.
Article34. Yu G, Mu H, Fang F, Zhou H, Li H, Wu Q, et al. LRP1B mutation associates with increased tumor mutation burden and inferior prognosis in liver hepatocellular carcinoma. Medicine (Baltimore). 2022; 101:e29763.
Article35. Brown LC, Tucker MD, Sedhom R, Schwartz EB, Zhu J, Kao C, et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J Immunother Cancer. 2021; 9:e001792.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Mixed adenoneuroendocrine carcinoma of the ampulla of Vater: Three case reports and a literature review
- Mixed Adenoneuroendocrine Gastric Carcinoma: A Case Report and Review of the Literature
- Gastric Mixed Adenoneuroendocrine Carcinoma with Revised Diagnosis after Retrospective Pathologic Review
- Multiregion Comprehensive Genomic Profiling of a Gastric Mixed Neuroendocrine-Nonneuroendocrine Neoplasm with Trilineage Differentiation
- Mixed Neuroendocrine-non-neuroendocrine Neoplasm of the Stomach that is Distributed in Depth on the Same Tumor: Inconsistent with the Definition of Mixed Adenoneuroendocrine Carcinoma in the 2010 World Health Organization Classification of Tumors of the Digestive System