Ann Lab Med.
2024 Nov;44(6):518-528. 10.3343/alm.2024.0039.
Detecting M-Protein via Mass Spectrometry and Affinity Beads: Enrichment With Mixed Kappa-Lambda Beads Enables Prompt Application in Clinical Laboratories
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea
- 3Digital OMICs Research Center, Korea Basic Science Institute, Cheongju, Korea
- 4College of Pharmacy, Chungnam National University, Daejeon, Korea
- 5Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
- 6Department of Laboratory Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 7Department of Laboratory Medicine, National Cancer Center, Goyang, Korea
- KMID: 2560798
- DOI: http://doi.org/10.3343/alm.2024.0039
Abstract
- Background
Detecting monoclonal protein (M-protein), a hallmark of plasma cell disorders, traditionally relies on methods such as protein electrophoresis, immune-electrophoresis, and immunofixation electrophoresis (IFE). Mass spectrometry (MS)-based methods, such as matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and electrospray ionization-quadrupole time-of-flight (ESI-qTOF) MS, have emerged as sensitive methods. We explored the M-protein-detection efficacies of different MS techniques.
Methods
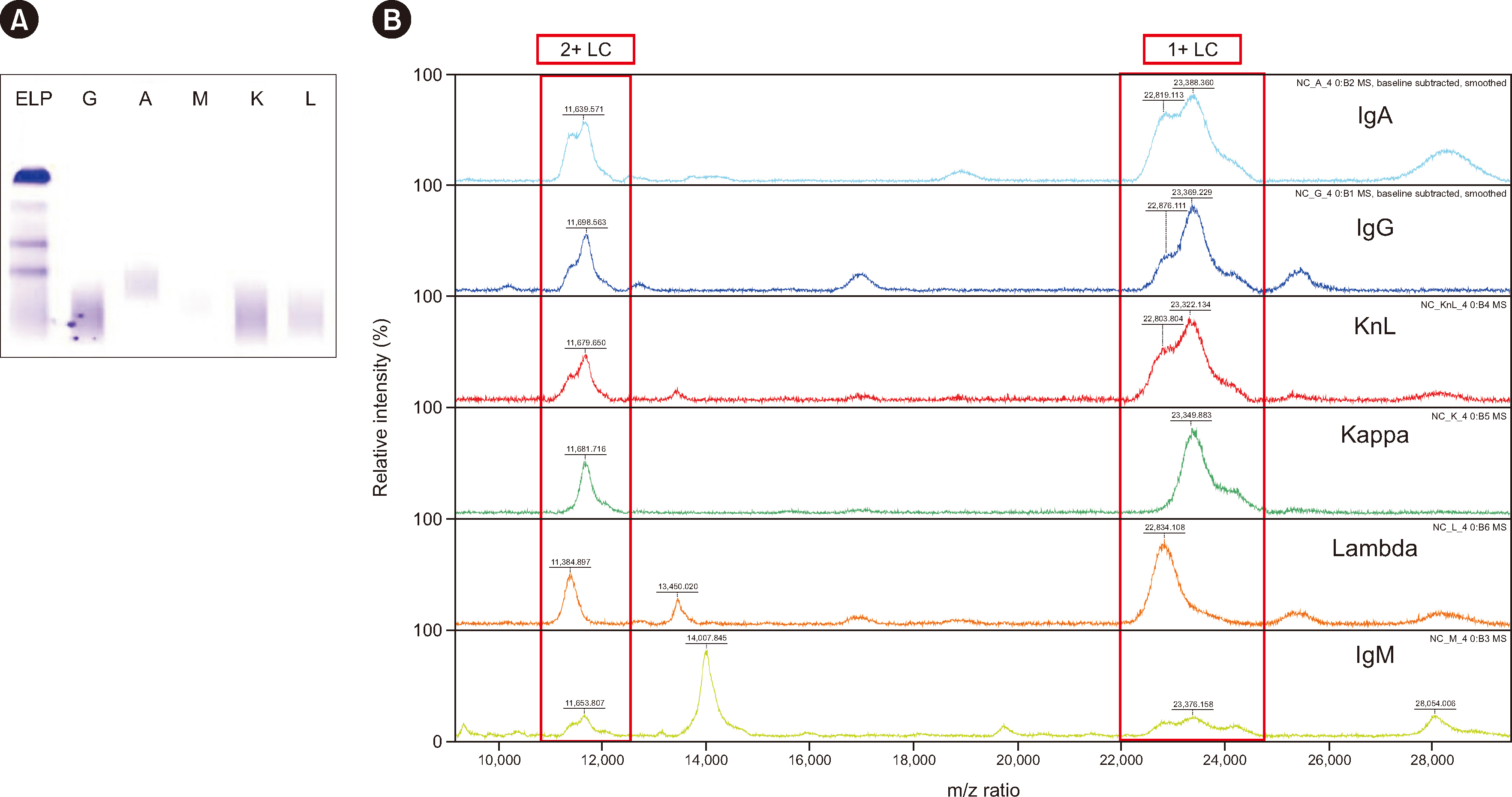
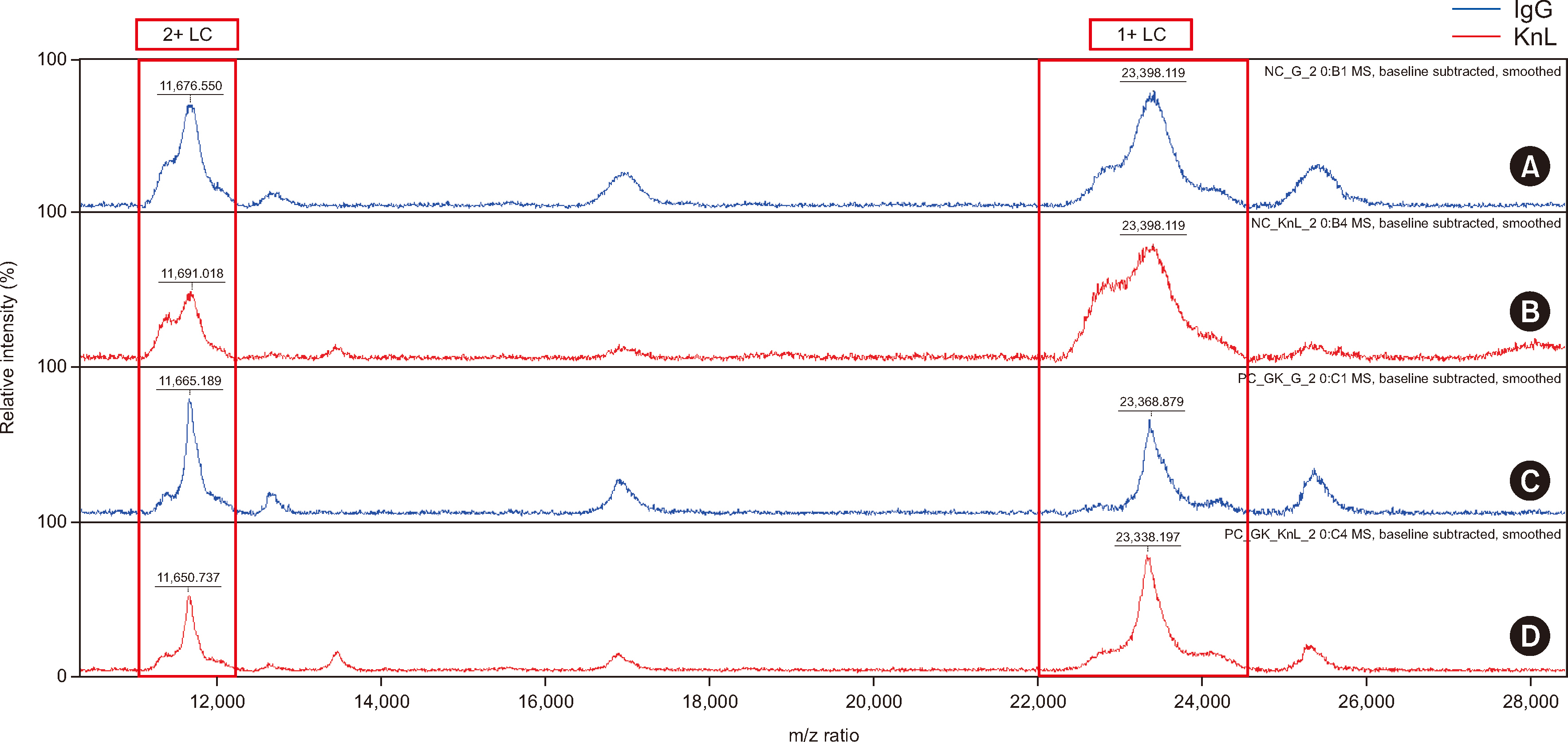
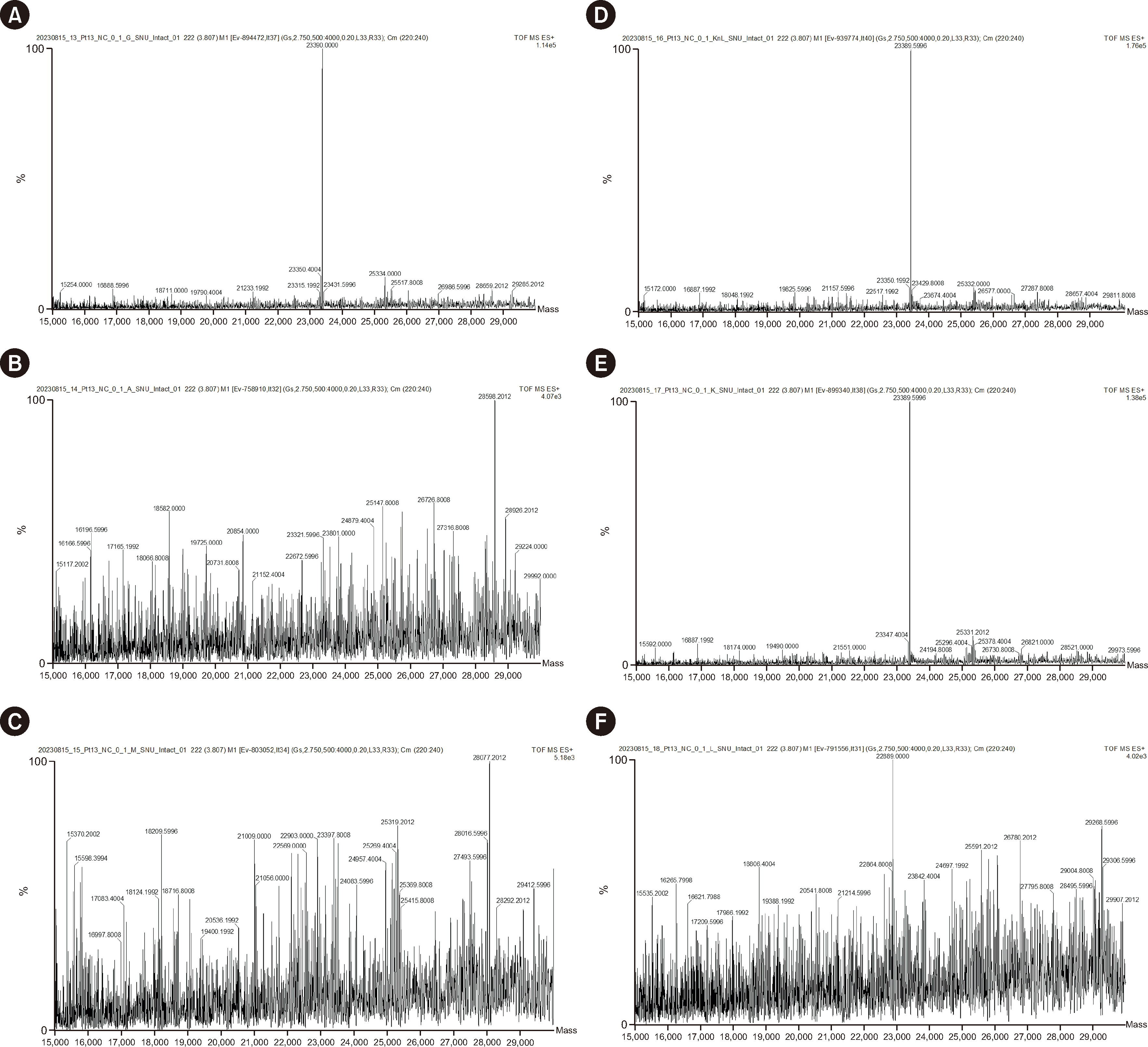
To isolate immunoglobulin and light chain proteins, six types of beads (IgG, IgA, IgM, kappa, lambda, and mixed kappa and lambda) were used to prepare samples along with CaptureSelect nanobody affinity beads (NBs). After purification, both MALDI-TOF MS and liquid chromatography coupled with Synapt G2 ESI-qTOF high-resolution MS analysis were performed. We purified 25 normal and 25 abnormal IFE samples using NBs and MALDI-TOF MS (NB-MALDI-TOF).
Results
Abnormal samples showed monoclonal peaks, whereas normal samples showed polyclonal peaks. The IgG and mixed kappa and lambda beads showed monoclonal peaks following the use of daratumumab (an IgG/kappa type of monoclonal antibody) with both MALDI-TOF and ESI-qTOF MS analysis. The limits of detection for MALDI-TOF MS and ESIqTOF MS were established as 0.1 g/dL and 0.025 g/dL, respectively. NB-MALDI-TOF and IFE exhibited comparable sensitivity and specificity (92% and 92%, respectively).
Conclusions
NBs for M-protein detection, particularly with mixed kappa-lambda beads, identified monoclonal peaks with both MALDI-TOF and ESI-qTOF analyses. Qualitative analysis using MALDI-TOF yielded results comparable with that of IFE. NB-MALDI-TOF might be used as an alternative method to replace conventional tests (such as IFE) to detect M-protein with high sensitivity.
Keyword
Figure
Reference
-
References
1. Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. 2014; International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15:e538–48. DOI: 10.1016/S1470-2045(14)70442-5. PMID: 25439696.2. International Myeloma Working Group. 2003; Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 121:749–57. DOI: 10.1046/j.1365-2141.2003.04355.x.3. Katzmann JA, Kyle RA, Benson J, Larson DR, Snyder MR, Lust JA, et al. 2009; Screening panels for detection of monoclonal gammopathies. Clin Chem. 55:1517–22. DOI: 10.1373/clinchem.2009.126664. PMID: 19520758. PMCID: PMC3773468.4. Vavricka SR, Burri E, Beglinger C, Degen L, Manz M. 2009; Serum protein electrophoresis: an underused but very useful test. Digestion. 79:203–10. DOI: 10.1159/000212077. PMID: 19365122.5. Mrosewski I, Urbank M. 2021; Identification of paraproteins via serum immunofixation or serum immunosubtraction and immunoturbidimetric quantitation of serum immunoglobulins in the laboratory testing for monoclonal gammopathies. Arch Pathol Lab Med. 145:1552–7. DOI: 10.5858/arpa.2020-0441-OA. PMID: 33635966.6. Dispenzieri A, Kyle R, Merlini G, Miguel JS, Ludwig H, Hajek R, et al. 2009; International Myeloma Working Group guidelines for serum-free light chain analysis in multiple myeloma and related disorders. Leukemia. 23:215–24. DOI: 10.1038/leu.2008.307. PMID: 19020545.7. Oliveros Conejero R, Pascual Usandizaga P, Garrido Chércoles A. 2020; Optimization of workflow and screening panels for the detection of malignant monoclonal gammopathies. Adv Lab Med. 1:20200042. DOI: 10.1515/almed-2020-0042. PMID: 37361501. PMCID: PMC10197466. PMID: 132a2c46435f44cb9221bd29608030d0.8. McCudden CR, Jacobs JFM, Keren D, Caillon H, Dejoie T, Andersen K. 2018; Recognition and management of common, rare, and novel serum protein electrophoresis and immunofixation interferences. Clin Biochem. 51:72–9. DOI: 10.1016/j.clinbiochem.2017.08.013. PMID: 28843491.9. Lim SM, Wijeratne N, Choy KW, Nguyen TTH, Setiawan L, Loh TP. 2024; A review of clinical guidelines, laboratory recommendations and external quality assurance programs for monoclonal gammopathy testing. Crit Rev Clin Lab Sci. 61:107–26. DOI: 10.1080/10408363.2023.2257306. PMID: 37776896.10. Bhole MV, Sadler R, Ramasamy K. 2014; Serum-free light-chain assay: clinical utility and limitations. Ann Clin Biochem. 51:528–42. DOI: 10.1177/0004563213518758. PMID: 24489083.11. Chapman JR, Thoren KL. 2020; Tracking of low disease burden in multiple myeloma: using mass spectrometry assays in peripheral blood. Best Pract Res Clin Haematol. 33:101142. DOI: 10.1016/j.beha.2020.101142. PMID: 32139008.12. Giles HV, Wechalekar A, Pratt G. 2022; The potential role of mass spectrometry for the identification and monitoring of patients with plasma cell disorders: where are we now and which questions remain unanswered? Br J Haematol. 198:641–53. DOI: 10.1111/bjh.18226. PMID: 35514140.13. Barnidge DR, Dasari S, Botz CM, Murray DH, Snyder MR, Katzmann JA, et al. 2014; Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J Proteome Res. 13:1419–27. DOI: 10.1021/pr400985k. PMID: 24467232.14. Milani P, Murray DL, Barnidge DR, Kohlhagen MC, Mills JR, Merlini G, et al. 2017; The utility of MASS-FIX to detect and monitor monoclonal proteins in the clinic. Am J Hematol. 92:772–9. DOI: 10.1002/ajh.24772. PMID: 28439985.15. Fatica EM, Martinez M, Ladwig PM, Murray JD, Kohlhagen MC, Kyle RA, et al. 2021; MALDI-TOF mass spectrometry can distinguish immunofixation bands of the same isotype as monoclonal or biclonal proteins. Clin Biochem. 97:67–73. DOI: 10.1016/j.clinbiochem.2021.08.001. PMID: 34384797.16. Kohlhagen MC, Barnidge DR, Mills JR, Stoner J, Gurtner KM, Liptac AM, et al. 2016; Screening method for M-proteins in serum using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin Chem. 62:1345–52. DOI: 10.1373/clinchem.2015.253781. PMID: 27515443.17. Kohlhagen M, Dasari S, Willrich M, Hetrick M, Netzel B, Dispenzieri A, et al. 2020; Automation and validation of a MALDI-TOF MS (Mass-Fix) replacement of immunofixation electrophoresis in the clinical lab. Clin Chem Lab Med. 59:155–63. DOI: 10.1515/cclm-2020-0581. PMID: 32745067.18. Ren Y, Shi Z, Zhang C, Han Y, Liu S, Hao P. 2022; Evaluation and minimization of over-alkylation in proteomic sample preparation. Int J Mass Spectrom. 481:116919. DOI: 10.1016/j.ijms.2022.116919.19. Müller T, Winter D. 2017; Systematic evaluation of protein reduction and alkylation reveals massive unspecific side effects by iodine-containing reagents. Mol Cell Proteomics. 16:1173–87. DOI: 10.1074/mcp.M116.064048. PMID: 28539326. PMCID: PMC5500753.20. Booth RA, McCudden CR, Balion CM, Blasutig IM, Bouhtiauy I, Rodriguez-Capote K, et al. 2018; Candidate recommendations for protein electrophoresis reporting from the Canadian Society of Clinical Chemists Monoclonal Gammopathy Working Group. Clin Biochem. 51:10–20. DOI: 10.1016/j.clinbiochem.2017.10.013. PMID: 29061378.21. Tang F, Malek E, Math S, Schmotzer CL, Beck RC. 2018; Interference of therapeutic monoclonal antibodies with routine serum protein electrophoresis and immunofixation in patients with myeloma: frequency and duration of detection of daratumumab and elotuzumab. Am J Clin Pathol. 150:121–9. DOI: 10.1093/ajcp/aqy037. PMID: 29901687.22. Rappold BA. 2022; Review of the use of liquid chromatography-tandem mass spectrometry in clinical laboratories: part I-development. Ann Lab Med. 42:121–40. DOI: 10.3343/alm.2022.42.2.121. PMID: 34635606. PMCID: PMC8548246.23. Bertamini L, D'Agostino M, Gay F. 2021; MRD assessment in multiple myeloma: progress and challenges. Curr Hematol Malig Rep. 16:162–71. DOI: 10.1007/s11899-021-00633-5. PMID: 33950462.24. Ladwig PM, Barnidge DR, Willrich MAV. 2017; Mass spectrometry approaches for identification and quantitation of therapeutic monoclonal antibodies in the clinical laboratory. Clin Vaccine Immunol. 24:e00545–16. DOI: 10.1128/CVI.00545-16. PMID: 28274937. PMCID: PMC5424237.25. Murray DL, Puig N, Kristinsson S, Usmani SZ, Dispenzieri A, Bianchi G, et al. 2021; Mass spectrometry for the evaluation of monoclonal proteins in multiple myeloma and related disorders: an International Myeloma Working Group Mass Spectrometry Committee Report. Blood Cancer J. 11:24. DOI: 10.1038/s41408-021-00408-4. PMID: 33563895. PMCID: PMC7873248. PMID: dee72de3162d43b8836c27ef0bcaf4e6.26. Keren DF, Schroeder L. 2016; Challenges of measuring monoclonal proteins in serum. Clin Chem Lab Med. 54:947–61. DOI: 10.1515/cclm-2015-0862. PMID: 26910744.27. McDonald Z, Taylor P, Liyasova M, Liu Q, Ma B. 2021; Mass spectrometry provides a highly sensitive noninvasive means of sequencing and tracking M-protein in the blood of multiple myeloma patients. J Proteome Res. 20:4176–85. DOI: 10.1021/acs.jproteome.0c01022. PMID: 34242034.28. Rappold BA. 2022; Review of the use of liquid chromatography-tandem mass spectrometry in clinical laboratories: part II-operations. Ann Lab Med. 42:531–57. DOI: 10.3343/alm.2022.42.5.531. PMID: 35470272. PMCID: PMC9057814.29. Liyasova M, McDonald Z, Taylor P, Gorospe K, Xu X, Yao C, et al. 2021; A personalized mass spectrometry-based assay to monitor M-protein in patients with multiple myeloma (EasyM). Clin Cancer Res. 27:5028–37. DOI: 10.1158/1078-0432.CCR-21-0649. PMID: 34210683. PMCID: PMC9401514.30. Murray DL. 2022; Bringing mass spectrometry into the care of patients with multiple myeloma. Int J Hematol. 115:790–8. DOI: 10.1007/s12185-022-03364-2. PMID: 35471500.31. Mills JR, Barnidge DR, Murray DL. 2015; Detecting monoclonal immunoglobulins in human serum using mass spectrometry. Methods. 81:56–65. DOI: 10.1016/j.ymeth.2015.04.020. PMID: 25916620.32. Goulet DR, Atkins WM. 2020; Considerations for the design of antibody-based therapeutics. J Pharm Sci. 109:74–103. DOI: 10.1016/j.xphs.2019.05.031. PMID: 31173761. PMCID: PMC6891151.33. Mills JR, Kohlhagen MC, Dasari S, Vanderboom PM, Kyle RA, Katzmann JA, et al. 2016; Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin Chem. 62:1334–44. DOI: 10.1373/clinchem.2015.253740. PMID: 27540026.34. Santockyte R, Puig O, Zheng N, Ouyang Z, Titsch C, Zhang YJ, et al. 2021; High-throughput therapeutic antibody interference-free high-resolution mass spectrometry assay for monitoring M-proteins in multiple myeloma. Anal Chem. 93:834–42. DOI: 10.1021/acs.analchem.0c03357. PMID: 33300779.35. Muccio S, Hirtz C, Descloux S, Fedeli O, Macé S, Lehmann S, et al. 2024; A sensitive high-resolution mass spectrometry method for quantifying intact M-protein light chains in patients with multiple myeloma. Clin Chim Acta. 552:117634. DOI: 10.1016/j.cca.2023.117634. PMID: 37980975.36. Sarto C, Intra J, Fania C, Brivio R, Brambilla P, Leoni V. 2021; Monoclonal free light chain detection and quantification: performances and limits of available laboratory assays. Clin Biochem. 95:28–33. DOI: 10.1016/j.clinbiochem.2021.05.006. PMID: 33991536.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of ingested multiple magnetic beads: the importance of suspicion based on medical history
- Elution profiles of metronidazole from calcium sulfate beads
- Effects of Different Sizes of Glass Beads on the Release of Sporocysts from Eimeria tenella Oocysts
- Two cases of intestinal obstruction due to ingestion of water beads
- A case of ingested water beads diagnosed with point-of-care ultrasound