J Pathol Transl Med.
2024 Sep;58(5):229-240. 10.4132/jptm.2024.07.13.
Artificial intelligence algorithm for neoplastic cell percentage estimation and its application to copy number variation in urinary tract cancer
- Affiliations
-
- 1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea
- KMID: 2559068
- DOI: http://doi.org/10.4132/jptm.2024.07.13
Abstract
- Background
Bladder cancer is characterized by frequent mutations, which provide potential therapeutic targets for most patients. The effectiveness of emerging personalized therapies depends on an accurate molecular diagnosis, for which the accurate estimation of the neoplastic cell percentage (NCP) is a crucial initial step. However, the established method for determining the NCP, manual counting by a pathologist, is time-consuming and not easily executable.
Methods
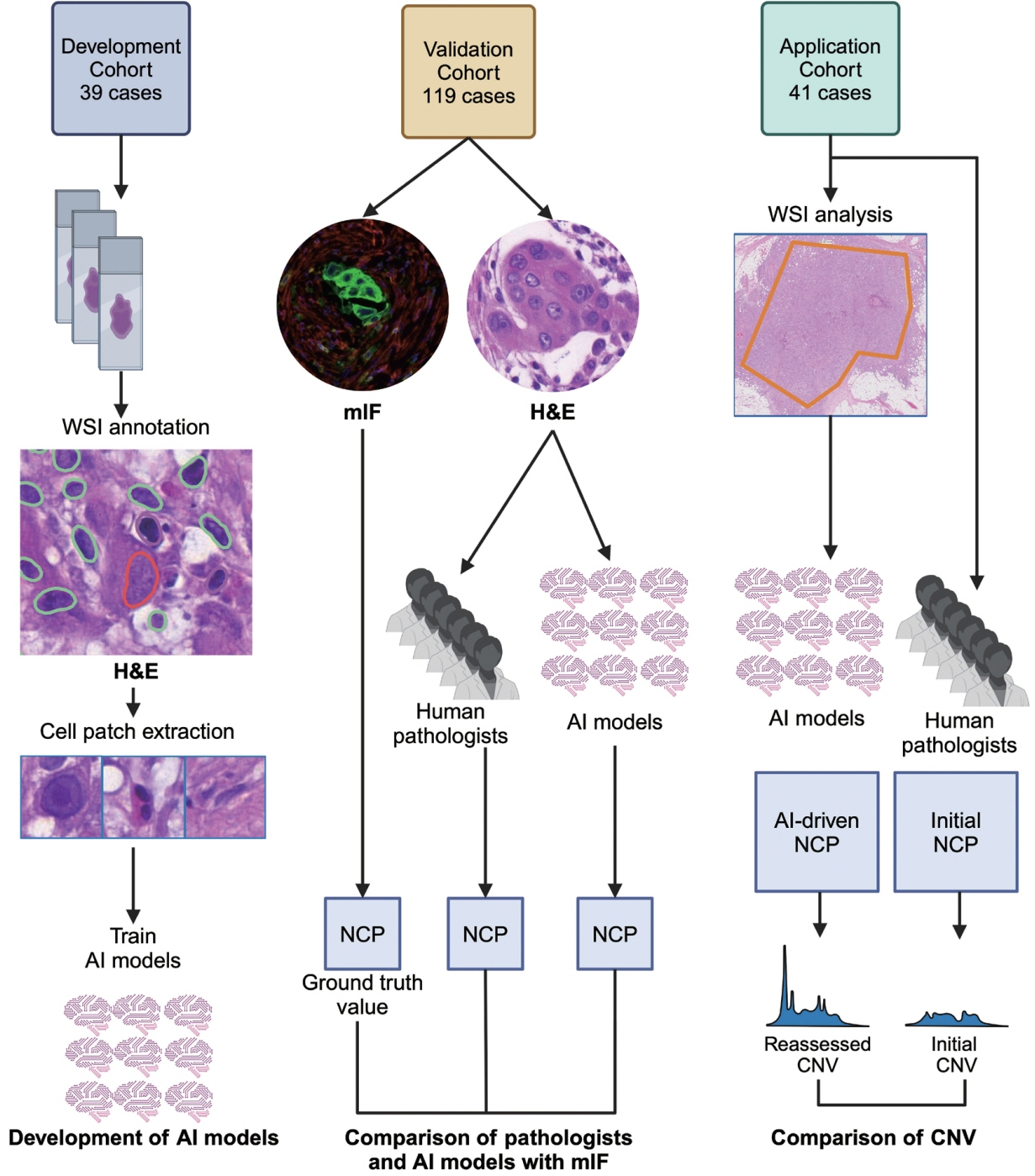
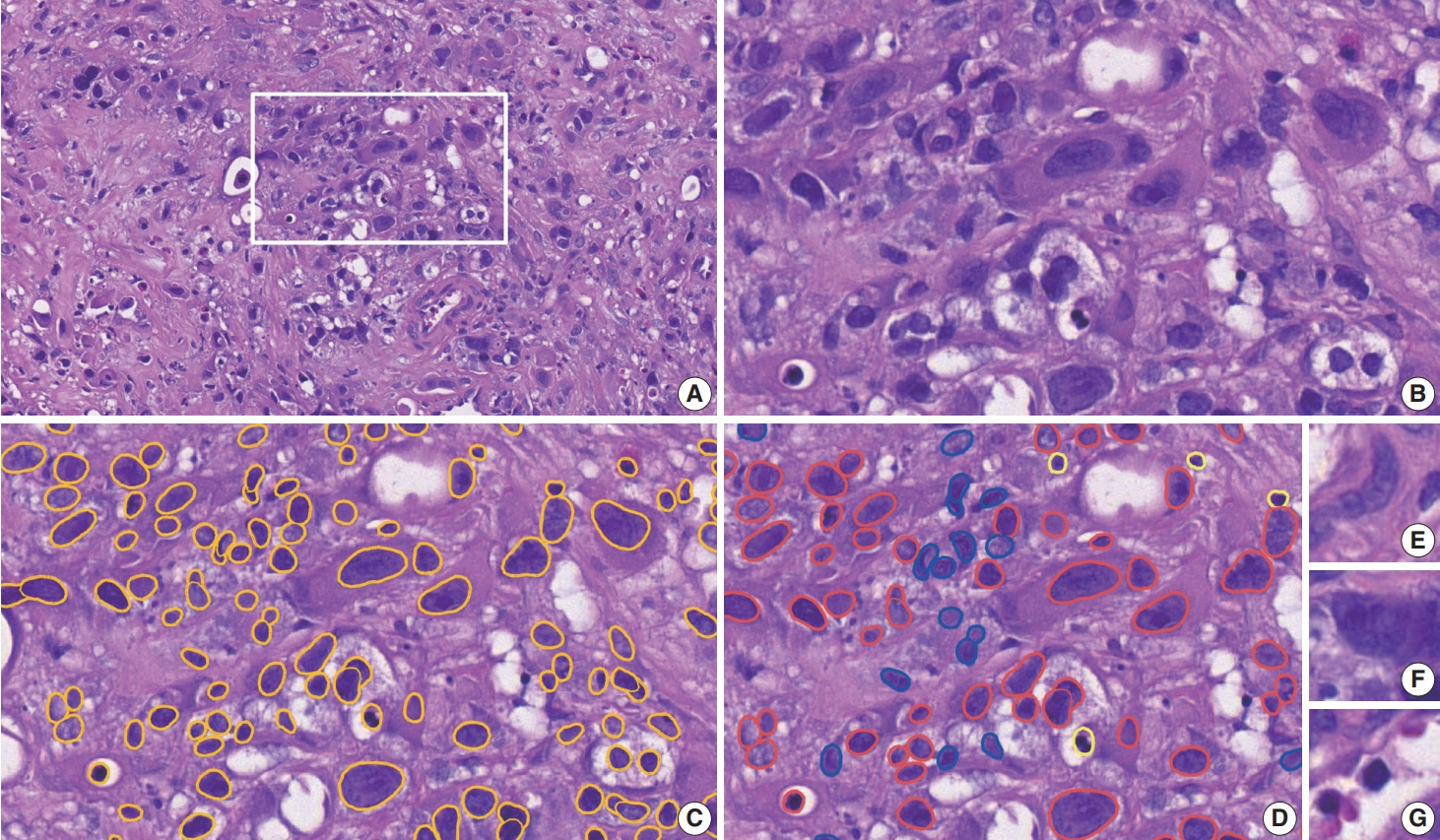
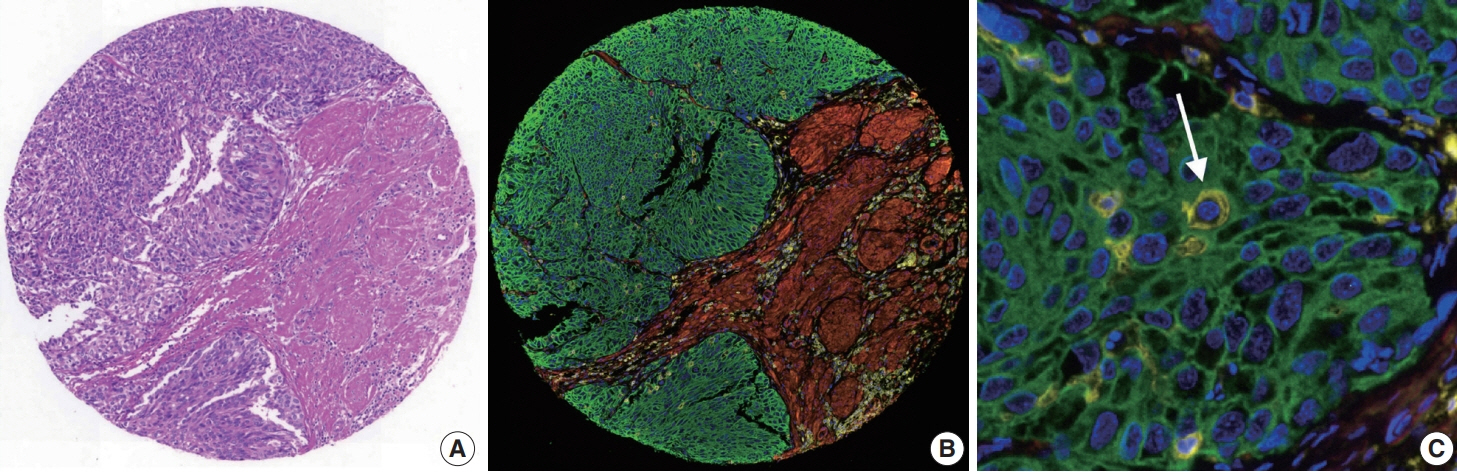
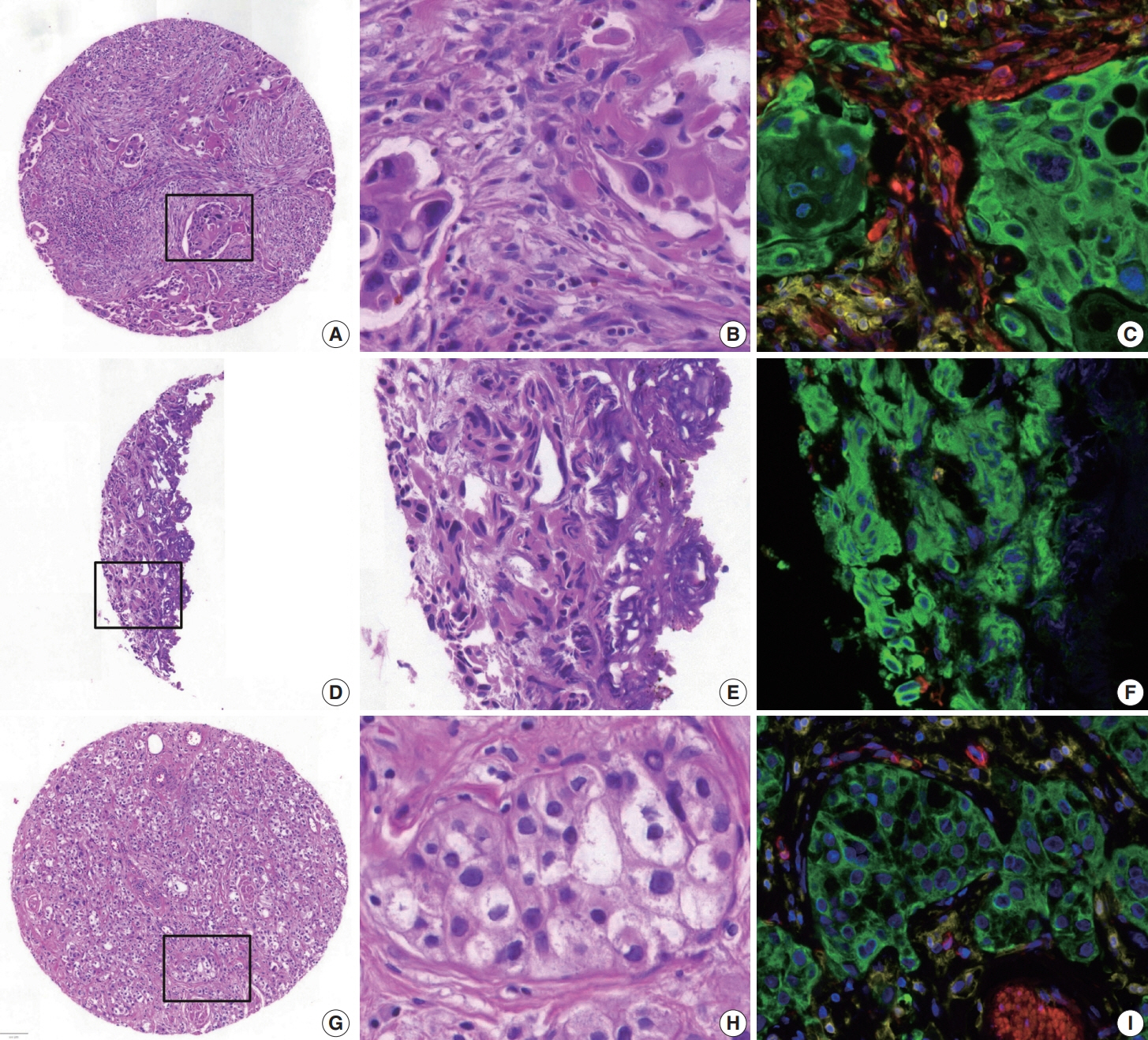
To address this, artificial intelligence (AI) models were developed to estimate the NCP using nine convolutional neural networks and the scanned images of 39 cases of urinary tract cancer. The performance of the AI models was compared to that of six pathologists for 119 cases in the validation cohort. The ground truth value was obtained through multiplexed immunofluorescence. The AI model was then applied to 41 cases in the application cohort that underwent next-generation sequencing testing, and its impact on the copy number variation (CNV) was analyzed.
Results
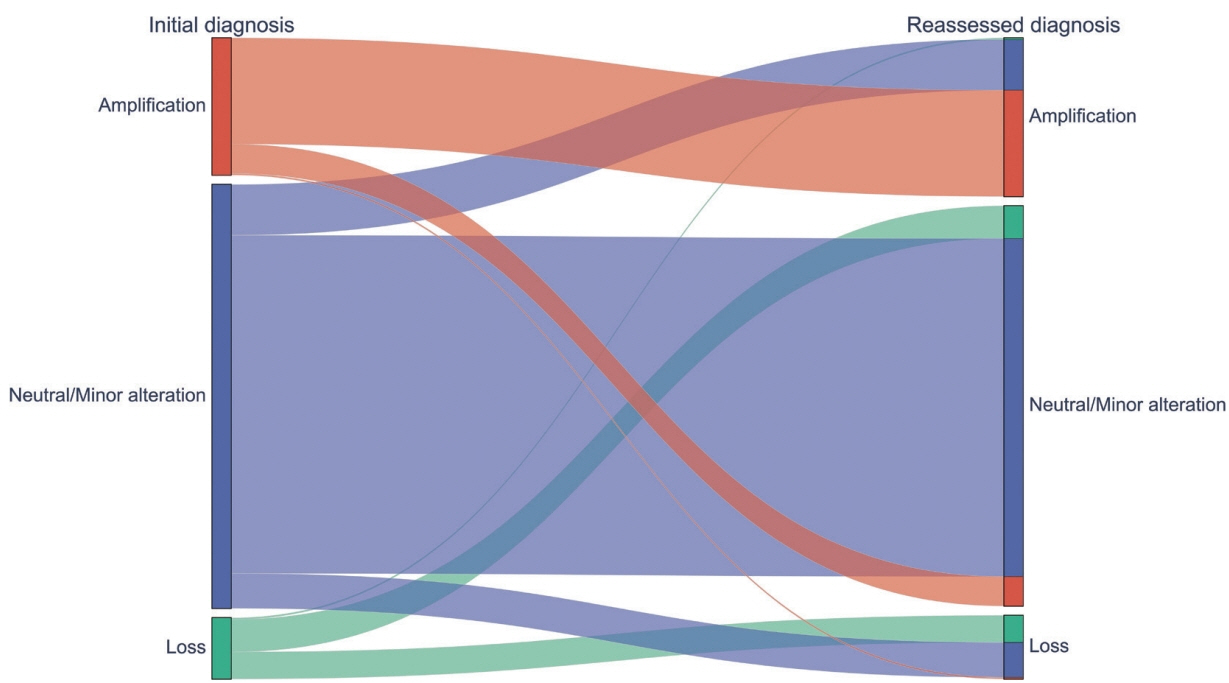
Each AI model demonstrated high reliability, with intraclass correlation coefficients (ICCs) ranging from 0.82 to 0.88. These values were comparable or better to those of pathologists, whose ICCs ranged from 0.78 to 0.91 in urothelial carcinoma cases, both with and without divergent differentiation/ subtypes. After applying AI-driven NCP, 190 CNV (24.2%) were reclassified with 66 (8.4%) and 78 (9.9%) moved to amplification and loss, respectively, from neutral/minor CNV. The neutral/minor CNV proportion decreased by 6%.
Conclusions
These results suggest that AI models could assist human pathologists in repetitive and cumbersome NCP calculations.
Figure
Reference
-
References
1. Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018; 174:1033.2. Loriot Y, Necchi A, Park SH, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019; 381:338–48.3. Sheng X, Yan X, Wang L, et al. Open-label, multicenter, phase II study of RC48-ADC, a HER2-targeting antibody-drug conjugate, in patients with locally advanced or metastatic urothelial carcinoma. Clin Cancer Res. 2021; 27:43–51.4. Patelli G, Zeppellini A, Spina F, et al. The evolving panorama of HER2-targeted treatments in metastatic urothelial cancer: a systematic review and future perspectives. Cancer Treat Rev. 2022; 104:102351.5. Zare F, Dow M, Monteleone N, Hosny A, Nabavi S. An evaluation of copy number variation detection tools for cancer using whole exome sequencing data. BMC Bioinformatics. 2017; 18:286.6. Smits AJ, Kummer JA, de Bruin PC, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014; 27:168–74.7. Dufraing K, van Krieken JH, De Hertogh G, et al. Neoplastic cell percentage estimation in tissue samples for molecular oncology: recommendations from a modified Delphi study. Histopathology. 2019; 75:312–9.8. Haider S, Tyekucheva S, Prandi D, et al. Systematic assessment of tumor purity and its clinical implications. JCO Precis Oncol. 2020; 4:PO.20.00016.9. Yadav VK, De S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogeneous tumor tissue samples. Brief Bioinform. 2015; 16:232–41.10. Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022; 35:23–32.11. Azimi V, Chang YH, Thibault G, et al. Breast cancer histopathology image analysis pipeline for tumor purity estimation. Proc IEEE Int Symp Biomed Imaging. 2017; 2017:1137–40.12. Lin S, Samsoondar JP, Bandari E, et al. Digital quantification of tumor cellularity as a novel prognostic feature of non-small cell lung carcinoma. Mod Pathol. 2023; 36:100055.13. Amin MB. Histological variants of urothelial carcinoma: diagnostic, therapeutic and prognostic implications. Mod Pathol. 2009; 22 Suppl 2:S96–118.14. Ahn J, Jin M, Song E, et al. Immune profiling of advanced thyroid cancers using fluorescent multiplex immunohistochemistry. Thyroid. 2021; 31:61–7.15. Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017; 7:16878.16. Schmidt U, Weigert M, Broaddus C, Myers G. Cell detection with star-convex polygons. In: Medical Image Computing and Computer Assisted Intervention - MICCAI 2018, Lecture Notes in Computer Science, Vol. 11071; 2018 Sep 16-20; Granada, Spain.17. Paszke A, Gross S, Massa F, et al. Pytorch: an imperative style, highperformance deep learning library. In: Advances in Neural Information Processing Systems 32, NeurIPS 2019; 2019 Dec 8-14; Vancouver, BC, Canada.18. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems, Vol. 1; 2012 Dec 3-6; Lake Tahoe, Nevada, CA, USA.19. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Preprint arXiv. at: https://doi.org/10.48550/arXiv.1409.1556 (2014).20. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, USA.21. Zagoruyko S, Komodakis N. Wide residual networks. Preprint arXiv. at: https://doi.org/10.48550/arXiv.1605.07146 (2016).22. Tan M, Le Q. EfficientNet: rethinking model scaling for convolutional neural networks. In: Proceedings of the 36th International Conference on Machine Learning (PMLR); 2019 Jun 9-15; Long Beach, CA, USA.23. Tan M, Le Q. Efficientnetv2: smaller models and faster training. In: Proceedings of the 38th International Conference on Machine Learning (PMLR); 2021 Jul 18-24; Virtual Event.24. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC. Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18-23; Salt Lake City, UT, USA.25. Howard A, Sandler M, Chen B, et al. Searching for MobileNetV3. In: Proceedings of the IEEE/CVF International Conference on Computer Vision; 2019 Oct 27-Nov 2; Seoul, Korea.26. Ma N, Zhang X, Zheng HT, Sun J. Shufflenet v2: practical guidelines for efficient CNN architecture design. In: Proceedings of the European Conference on Computer Vision (ECCV 2018); 2018 Sep 8-14; Munich, Germany.27. Kingma DP, Ba J. Adam: a method for stochastic optimization. Preprint arXiv. at: https://doi.org/10.48550/arXiv.1412.6980 (2014).28. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979; 86:420–8.29. Robert ME, Ruschoff J, Jasani B, et al. High interobserver variability among pathologists using combined positive score to evaluate PDL1 expression in gastric, gastroesophageal junction, and esophageal adenocarcinoma. Mod Pathol. 2023; 36:100154.30. Cicchetti DV. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess. 1994; 6:284–90.31. Kim JE, Chun SM, Hong YS, et al. Mutation burden and I index for detection of microsatellite instability in colorectal cancer by targeted next-generation sequencing. J Mol Diagn. 2019; 21:241–50.32. Kim M, Lee C, Hong J, et al. Validation and clinical application of ONCOaccuPanel for targeted next-generation sequencing of solid tumors. Cancer Res Treat. 2023; 55:429–41.33. Oh JH, Sung CO, Kim HD, Chun SM, Kim J. BRCA-mutated gastric adenocarcinomas are associated with chromosomal instability and responsiveness to platinum-based chemotherapy. J Pathol Transl Med. 2023; 57:323–31.34. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016; 12:e1004873.35. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations In cancer. Nucleic Acids Res. 2019; 47:D941–7.36. Zhao M, Zhao Z. Concordance of copy number loss and down-regulation of tumor suppressor genes: a pan-cancer study. BMC Genomics. 2016; 17 Suppl 7:532.37. Suehnholz SP, Nissan MH, Zhang H, et al. Quantifying the expanding landscape of clinical actionability for patients with cancer. Cancer Discov. 2024; 14:49–65.38. Homeyer A, Geissler C, Schwen LO, et al. Recommendations on compiling test datasets for evaluating artificial intelligence solutions in pathology. Mod Pathol. 2022; 35:1759–69.39. Lhermitte B, Egele C, Weingertner N, et al. Adequately defining tumor cell proportion in tissue samples for molecular testing improves interobserver reproducibility of its assessment. Virchows Arch. 2017; 470:21–7.40. Sakamoto T, Furukawa T, Pham HH, et al. A collaborative workflow between pathologists and deep learning for the evaluation of tumour cellularity in lung adenocarcinoma. Histopathology. 2022; 81:758–69.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Latest Trends in the Use of Deep Learning in Radiology Illustrated Through the Stages of Deep Learning Algorithm Development
- Application of artificial intelligence for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging
- Development and Evaluation of Urolithiasis Detection Technology Based on a Multimethod Algorithm
- Role of artificial intelligence in diagnosing Barrett’s esophagus-related neoplasia
- Application of Endoscopic Ultrasound-based Artificial Intelligence in Diagnosis of Pancreatic Malignancies