Diabetes Metab J.
2024 Sep;48(5):821-836. 10.4093/dmj.2024.0382.
Systems Biology of Human Microbiome for the Prediction of Personal Glycaemic Response
- Affiliations
-
- 1School of Life Sciences, Gwangju Institute of Science and Technology, Gwangju, Korea
- 2Division of Endocrinology and Metabolism, Department of Internal Medicine, Bucheon St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Bucheon, Korea
- KMID: 2558994
- DOI: http://doi.org/10.4093/dmj.2024.0382
Abstract
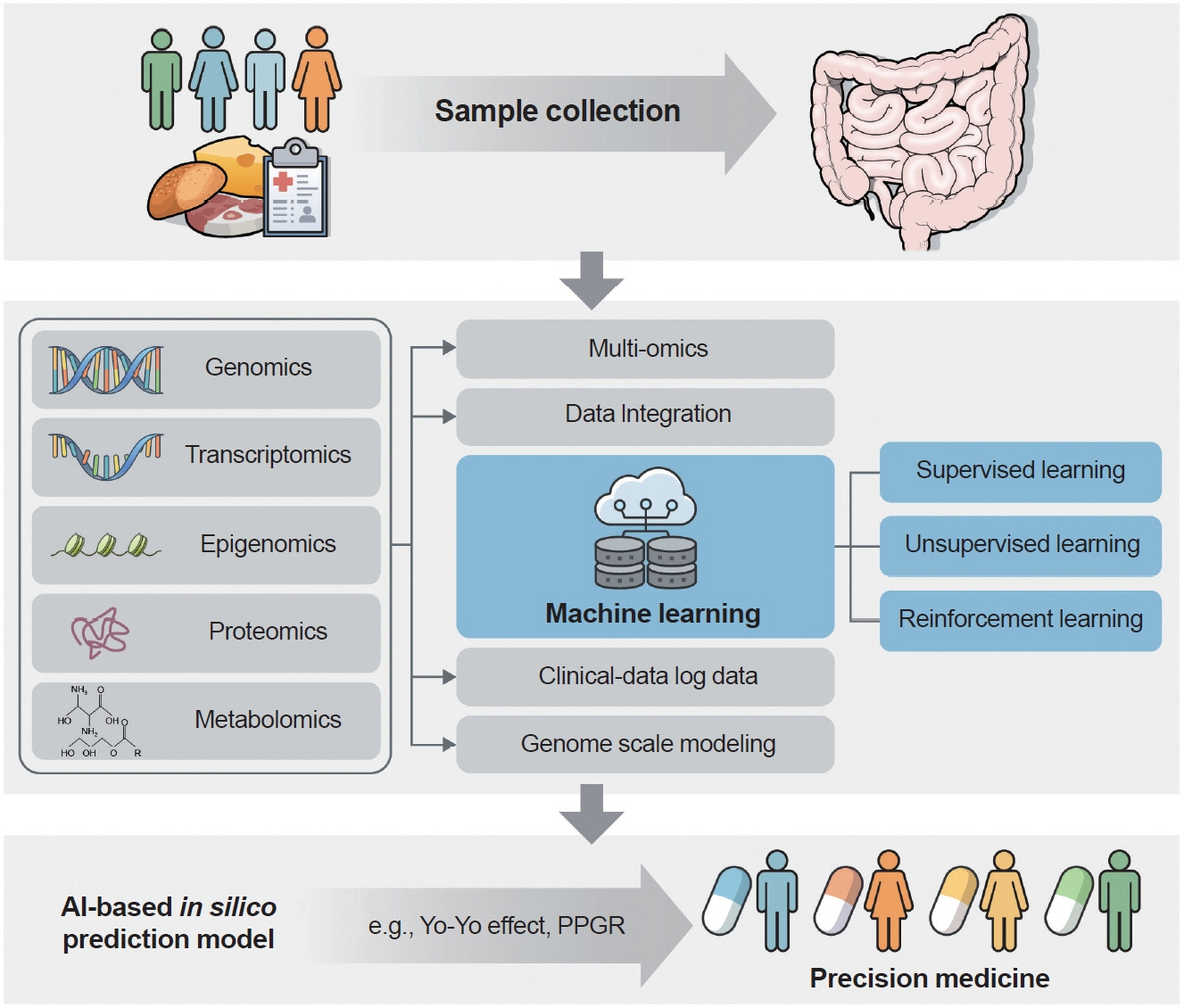
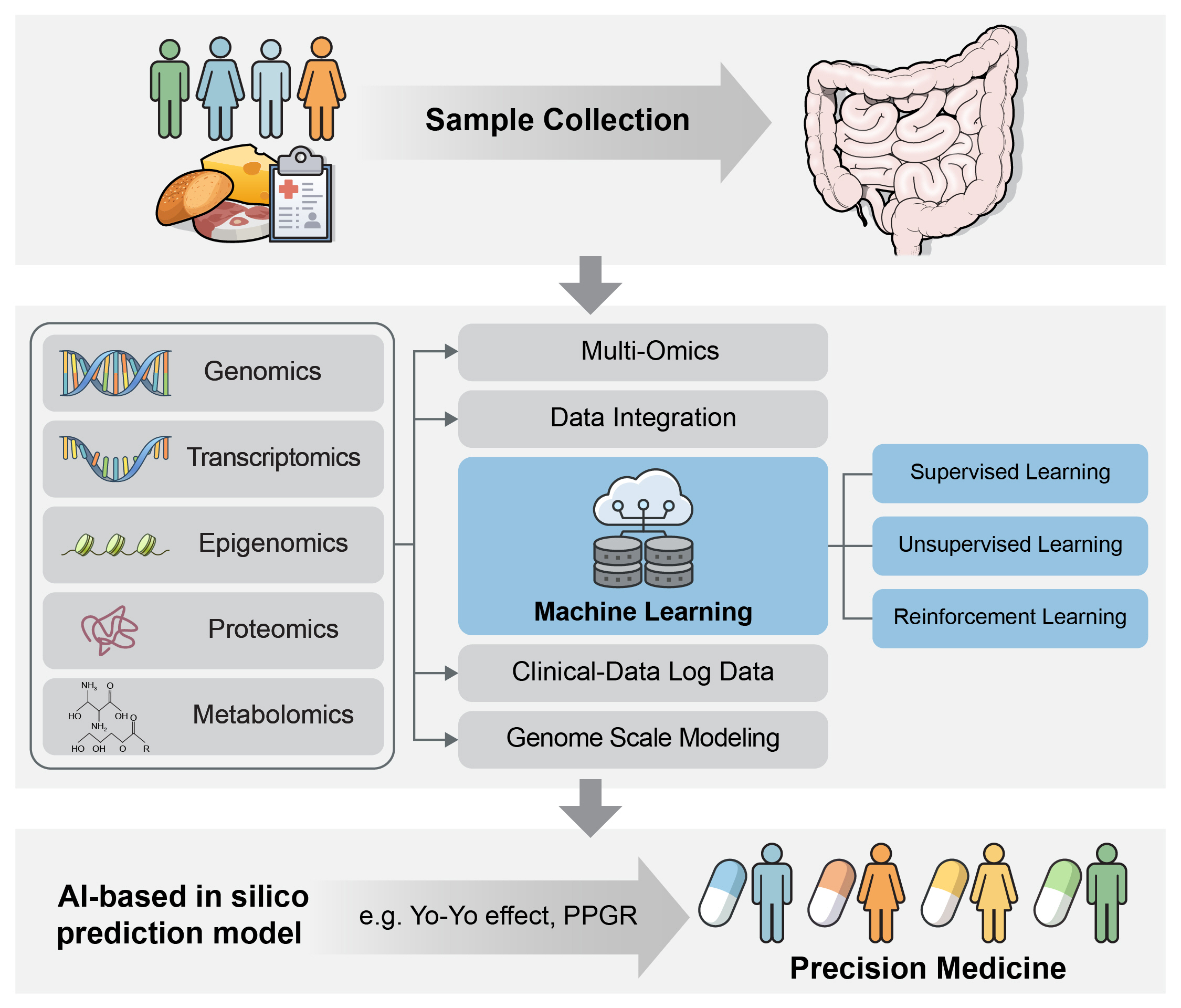
- The human gut microbiota is increasingly recognized as a pivotal factor in diabetes management, playing a significant role in the body’s response to treatment. However, it is important to understand that long-term usage of medicines like metformin and other diabetic treatments can result in problems, gastrointestinal discomfort, and dysbiosis of the gut flora. Advanced sequencing technologies have improved our understanding of the gut microbiome’s role in diabetes, uncovering complex interactions between microbial composition and metabolic health. We explore how the gut microbiota affects glucose metabolism and insulin sensitivity by examining a variety of -omics data, including genomics, transcriptomics, epigenomics, proteomics, metabolomics, and metagenomics. Machine learning algorithms and genome-scale modeling are now being applied to find microbiological biomarkers associated with diabetes risk, predicted disease progression, and guide customized therapy. This study holds promise for specialized diabetic therapy. Despite significant advances, some concerns remain unanswered, including understanding the complex relationship between diabetes etiology and gut microbiota, as well as developing user-friendly technological innovations. This mini-review explores the relationship between multiomics, precision medicine, and machine learning to improve our understanding of the gut microbiome’s function in diabetes. In the era of precision medicine, the ultimate goal is to improve patient outcomes through personalized treatments.
Figure
Reference
-
1. Magliano DJ, Boyko EJ. IDF diabetes atlas. Brussels: International Diabetes Federation;2022.2. Morrish NJ, Wang SL, Stevens LK, Fuller JH, Keen H. Mortality and causes of death in the WHO Multinational Study of Vascular Disease in Diabetes. Diabetologia. 2001; 44 Suppl 2:S14–21.
Article3. Hu FB, Satija A, Manson JE. Curbing the diabetes pandemic: the need for global policy solutions. JAMA. 2015; 313:2319–20.4. Jin Q, Ma RC. Metabolomics in diabetes and diabetic complications: insights from epidemiological studies. Cells. 2021; 10:2832.
Article5. Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019; 26:666–79.
Article6. Mitro SD, Liu J, Jaacks LM, Fleisch AF, Williams PL, Knowler WC, et al. Per- and polyfluoroalkyl substance plasma concentrations and metabolomic markers of type 2 diabetes in the Diabetes Prevention Program trial. Int J Hyg Environ Health. 2021; 232:113680.
Article7. Merino J, Leong A, Liu CT, Porneala B, Walford GA, von Grotthuss M, et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia. 2018; 61:1315–24.
Article8. Lu Y, Wang Y, Ong CN, Subramaniam T, Choi HW, Yuan JM, et al. Metabolic signatures and risk of type 2 diabetes in a Chinese population: an untargeted metabolomics study using both LC-MS and GC-MS. Diabetologia. 2016; 59:2349–59.
Article9. Morze J, Wittenbecher C, Schwingshackl L, Danielewicz A, Rynkiewicz A, Hu FB, et al. Metabolomics and type 2 diabetes risk: an updated systematic review and meta-analysis of prospective cohort studies. Diabetes Care. 2022; 45:1013–24.
Article10. Razquin C, Toledo E, Clish CB, Ruiz-Canela M, Dennis C, Corella D, et al. Plasma lipidomic profiling and risk of type 2 diabetes in the PREDIMED trial. Diabetes Care. 2018; 41:2617–24.
Article11. Yoo JY, Groer M, Dutra SV, Sarkar A, McSkimming DI. Gut microbiota and immune system interactions. Microorganisms. 2020; 8:1587.
Article12. Prada M, Wittenbecher C, Eichelmann F, Wernitz A, Drouin-Chartier JP, Schulze MB. Association of the odd-chain fatty acid content in lipid groups with type 2 diabetes risk: a targeted analysis of lipidomics data in the EPIC-Potsdam cohort. Clin Nutr. 2021; 40:4988–99.
Article13. Suvitaival T, Bondia-Pons I, Yetukuri L, Poho P, Nolan JJ, Hyotylainen T, et al. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism. 2018; 78:1–12.
Article14. Lee HS, Xu T, Lee Y, Kim NH, Kim YJ, Kim JN, et al. Identification of putative biomarkers for type 2 diabetes using metabolomics in the Korea Association REsource (KARE) cohort. Metabolomics. 2016; 12:178.
Article15. Mamtani M, Kulkarni H, Wong G, Weir JM, Barlow CK, Dyer TD, et al. Lipidomic risk score independently and cost-effectively predicts risk of future type 2 diabetes: results from diverse cohorts. Lipids Health Dis. 2016; 15:67.
Article16. Menni C, Fauman E, Erte I, Perry JR, Kastenmuller G, Shin SY, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013; 62:4270–6.
Article17. Sundsten T, Ortsater H. Proteomics in diabetes research. Mol Cell Endocrinol. 2009; 297:93–103.
Article18. Wang N, Zhu F, Chen L, Chen K. Proteomics, metabolomics and metagenomics for type 2 diabetes and its complications. Life Sci. 2018; 212:194–202.
Article19. Wu W, Feng J, Jiang D, Zhou X, Jiang Q, Cai M, et al. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine. Sci Rep. 2017; 7:41606.
Article20. Wagman AS, Johnson KW, Bussiere DE. Discovery and development of GSK3 inhibitors for the treatment of type 2 diabetes. Curr Pharm Des. 2004; 10:1105–37.
Article21. He RJ, Yu ZH, Zhang RY, Zhang ZY. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin. 2014; 35:1227–46.
Article22. Blaslov K, Bulum T, Zibar K, Duvnjak L. Relationship between adiponectin level, insulin sensitivity, and metabolic syndrome in type 1 diabetic patients. Int J Endocrinol. 2013; 2013:535906.
Article23. Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012; 13:383–96.
Article24. Kurauti MA, Costa-Junior JM, Ferreira SM, Santos GJ, Sponton CH, Carneiro EM, et al. Interleukin-6 increases the expression and activity of insulin-degrading enzyme. Sci Rep. 2017; 7:46750.
Article25. Huang C, Lin MZ, Cheng D, Braet F, Pollock CA, Chen XM. KCa3.1 mediates dysfunction of tubular autophagy in diabetic kidneys via PI3k/Akt/mTOR signaling pathways. Sci Rep. 2016; 6:23884.
Article26. Awata T, Neda T, Iizuka H, Kurihara S, Ohkubo T, Takata N, et al. Endothelial nitric oxide synthase gene is associated with diabetic macular edema in type 2 diabetes. Diabetes Care. 2004; 27:2184–90.
Article27. Wu SY, Wang GF, Liu ZQ, Rao JJ, Lu L, Xu W, et al. Effect of geniposide, a hypoglycemic glucoside, on hepatic regulating enzymes in diabetic mice induced by a high-fat diet and streptozotocin. Acta Pharmacol Sin. 2009; 30:202–8.
Article28. Li W, Yang X, Zheng T, Xing S, Wu Y, Bian F, et al. TNF-α stimulates endothelial palmitic acid transcytosis and promotes insulin resistance. Sci Rep. 2017; 7:44659.
Article29. Sun Y, Liu S, Ferguson S, Wang L, Klepcyk P, Yun JS, et al. Phosphoenolpyruvate carboxykinase overexpression selectively attenuates insulin signaling and hepatic insulin sensitivity in transgenic mice. J Biol Chem. 2002; 277:23301–7.
Article30. Celi FS, Negri C, Tanner K, Raben N, De Pablo F, Rovira A, et al. Molecular scanning for mutations in the insulin receptor substrate-1 (IRS-1) gene in Mexican Americans with type 2 diabetes mellitus. Diabetes Metab Res Rev. 2000; 16:370–7.
Article31. Tenenbaum A, Fisman EZ, Motro M. Metabolic syndrome and type 2 diabetes mellitus: focus on peroxisome proliferator activated receptors (PPAR). Cardiovasc Diabetol. 2003; 2:4.
Article32. Wan Y, Bao X, Huang J, Zhang X, Liu W, Cui Q, et al. Novel GLP-1 analog supaglutide reduces HFD-induced obesity associated with increased Ucp-1 in white adipose tissue in mice. Front Physiol. 2017; 8:294.
Article33. Treadway JL, Mendys P, Hoover DJ. Glycogen phosphorylase inhibitors for treatment of type 2 diabetes mellitus. Expert Opin Investig Drugs. 2001; 10:439–54.
Article34. Hogan JC, Stephens JM. The regulation of fatty acid synthase by STAT5A. Diabetes. 2005; 54:1968–75.
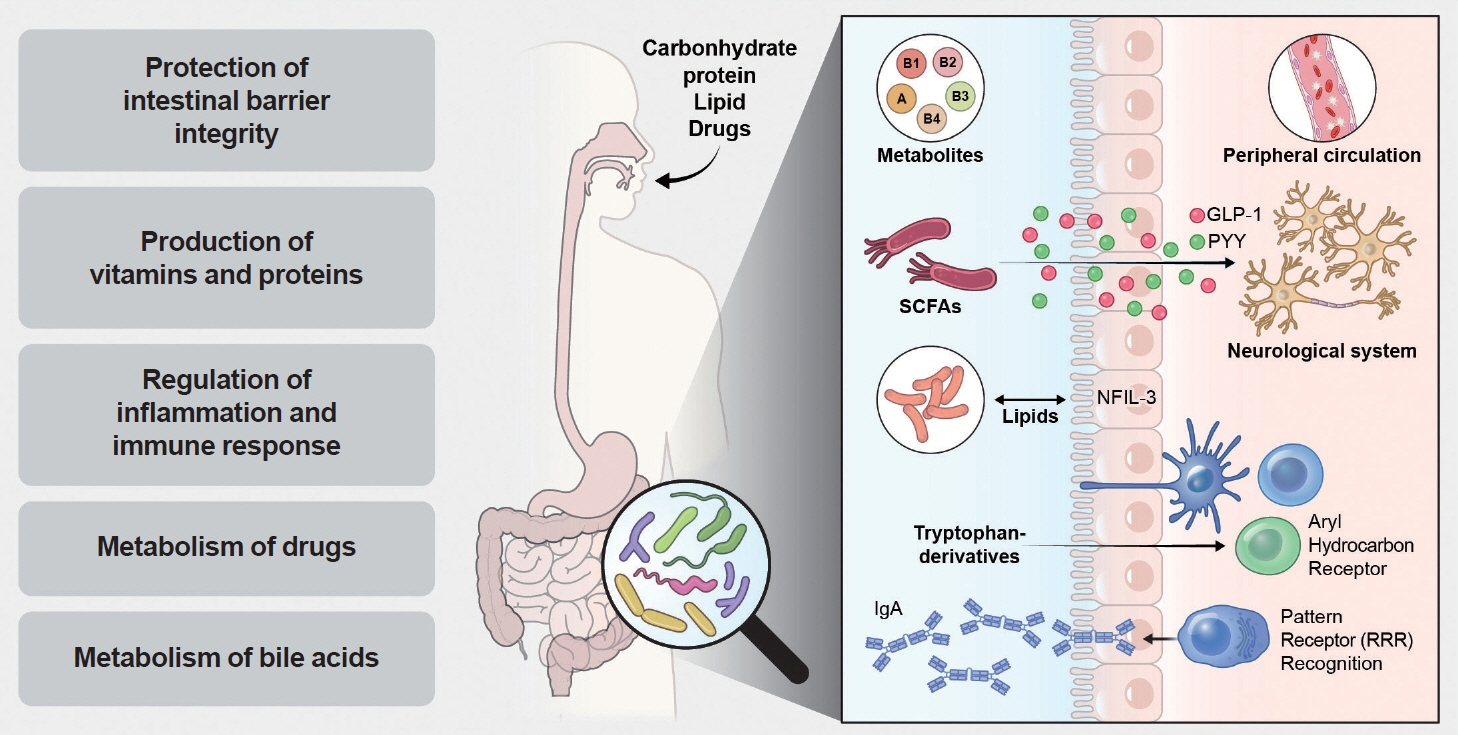
Article35. Herrema H, Niess JH. Intestinal microbial metabolites in human metabolism and type 2 diabetes. Diabetologia. 2020; 63:2533–47.
Article36. Dash S, Syed YA, Khan MR. Understanding the role of the gut microbiome in brain development and its association with neurodevelopmental psychiatric disorders. Front Cell Dev Biol. 2022; 10:880544.
Article37. Thaiss CA, Shapiro H, Elinav E. Post-dieting weight gain: the role of persistent microbiome changes. Future Microbiol. 2017; 12:555–9.
Article38. Cox TO, Lundgren P, Nath K, Thaiss CA. Metabolic control by the microbiome. Genome Med. 2022; 14:80.
Article39. Crudele L, Gadaleta RM, Cariello M, Moschetta A. Gut microbiota in the pathogenesis and therapeutic approaches of diabetes. EBioMedicine. 2023; 97:104821.
Article40. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019; 568:499–504.
Article41. Chen LX, Anantharaman K, Shaiber A, Eren AM, Banfield JF. Accurate and complete genomes from metagenomes. Genome Res. 2020; 30:315–33.
Article42. Bhattacharya T, Ghosh TS, Mande SS. Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS One. 2015; 10:e0142038.
Article43. Ghosh TS, Gupta SS, Nair GB, Mande SS. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS One. 2013; 8:e83823.
Article44. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007; 449:804–10.
Article45. Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014; 16:276–89.46. Navas-Molina JA, Hyde ER, Sanders J, Knight R. The microbiome and big data. Curr Opin Syst Biol. 2017; 4:92–6.
Article47. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015; 163:1079–94.
Article48. Song J, Oh TJ, Song Y. Individual postprandial glycemic responses to meal types by different carbohydrate levels and their associations with glycemic variability using continuous glucose monitoring. Nutrients. 2023; 15:3571.
Article49. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenomewide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490:55–60.
Article50. Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015; 26:493–501.
Article51. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3:289–306.
Article52. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004; 101:15718–23.
Article53. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–31.
Article54. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013; 341:1241214.
Article55. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018; 555:210–5.
Article56. Dekkers KF, Sayols-Baixeras S, Baldanzi G, Nowak C, Hammar U, Nguyen D, et al. An online atlas of human plasma metabolite signatures of gut microbiome composition. Nat Commun. 2022; 13:5370.
Article57. Chen L, Zhernakova DV, Kurilshikov A, Andreu-Sanchez S, Wang D, Augustijn HE, et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat Med. 2022; 28:2333–43.
Article58. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016; 16:341–52.
Article59. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013; 54:2325–40.
Article60. van der Hee B, Wells JM. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021; 29:700–12.
Article61. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003; 278:11312–9.
Article62. Tazoe H, Otomo Y, Karaki S, Kato I, Fukami Y, Terasaki M, et al. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009; 30:149–56.
Article63. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009; 461:1282–6.
Article64. Pluznick JL. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. 2016; 90:1191–8.
Article65. Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A. 2011; 108:8030–5.
Article66. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021; 19:55–71.
Article67. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012; 61:364–71.
Article68. Wei YH, Ma X, Zhao JC, Wang XQ, Gao CQ. Succinate metabolism and its regulation of host-microbe interactions. Gut Microbes. 2023; 15:2190300.
Article69. Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017; 21:208–19.
Article70. Ternes D, Tsenkova M, Pozdeev VI, Meyers M, Koncina E, Atatri S, et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat Metab. 2022; 4:458–75.
Article71. Xu W, Xue W, Zhou Z, Wang J, Qi H, Sun S, et al. Formate might be a novel potential serum metabolic biomarker for type 2 diabetic peripheral neuropathy. Diabetes Metab Syndr Obes. 2023; 16:3147–60.
Article72. Wang SP, Rubio LA, Duncan SH, Donachie GE, Holtrop G, Lo G, et al. Pivotal roles for pH, lactate, and lactate-utilizing bacteria in the stability of a human colonic microbial ecosystem. mSystems. 2020; 5:e00645–20.
Article73. Lee YS, Kim TY, Kim Y, Lee SH, Kim S, Kang SW, et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe. 2018; 24:833–46.
Article74. Zhang Q, Hu N. Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Metab Syndr Obes. 2020; 13:5003–14.75. Olokoba AB, Obateru OA, Olokoba LB. Type 2 diabetes mellitus: a review of current trends. Oman Med J. 2012; 27:269–73.
Article76. Harsch IA, Konturek PC. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into “old” diseases. Med Sci (Basel). 2018; 6:32.
Article77. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018; 359:1151–6.
Article78. Eisenberg Y, Dugas LR, Akbar A, Reddivari B, Layden BT, Barengolts E. Oxytocin is lower in African American men with diabetes and associates with psycho-social and metabolic health factors. PLoS One. 2018; 13:e0190301.
Article79. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020; 51:102590.
Article80. He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018; 24:1532–5.
Article81. Wang Y, Luo X, Mao X, Tao Y, Ran X, Zhao H, et al. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS One. 2017; 12:e0172774.
Article82. Yamashita H, Maruta H, Jozuka M, Kimura R, Iwabuchi H, Yamato M, et al. Effects of acetate on lipid metabolism in muscles and adipose tissues of type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biosci Biotechnol Biochem. 2009; 73:570–6.
Article83. den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 2015; 64:2398–408.
Article84. Yamashita H, Fujisawa K, Ito E, Idei S, Kawaguchi N, Kimoto M, et al. Improvement of obesity and glucose tolerance by acetate in type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biosci Biotechnol Biochem. 2007; 71:1236–43.
Article85. Kondo T, Kishi M, Fushimi T, Kaga T. Acetic acid upregulates the expression of genes for fatty acid oxidation enzymes in liver to suppress body fat accumulation. J Agric Food Chem. 2009; 57:5982–6.
Article86. Sahuri-Arisoylu M, Brody LP, Parkinson JR, Parkes H, Navaratnam N, Miller AD, et al. Reprogramming of hepatic fat accumulation and ‘browning’ of adipose tissue by the short-chain fatty acid acetate. Int J Obes (Lond). 2016; 40:955–63.
Article87. Ge H, Li X, Weiszmann J, Wang P, Baribault H, Chen JL, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008; 149:4519–26.
Article88. Hong YH, Nishimura Y, Hishikawa D, Tsuzuki H, Miyahara H, Gotoh C, et al. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005; 146:5092–9.
Article89. Jocken JW, Gonzalez Hernandez MA, Hoebers NT, van der Beek CM, Essers YP, Blaak EE, et al. Short-chain fatty acids differentially affect intracellular lipolysis in a human white adipocyte model. Front Endocrinol (Lausanne). 2017; 8:372.
Article90. Hanatani S, Motoshima H, Takaki Y, Kawasaki S, Igata M, Matsumura T, et al. Acetate alters expression of genes involved in beige adipogenesis in 3T3-L1 cells and obese KK-Ay mice. J Clin Biochem Nutr. 2016; 59:207–14.
Article91. Canfora EE, van der Beek CM, Jocken JW, Goossens GH, Holst JJ, Olde Damink SW, et al. Colonic infusions of short-chain fatty acid mixtures promote energy metabolism in overweight/obese men: a randomized crossover trial. Sci Rep. 2017; 7:2360.
Article92. van der Beek CM, Canfora EE, Lenaerts K, Troost FJ, Damink SW, Holst JJ, et al. Distal, not proximal, colonic acetate infusions promote fat oxidation and improve metabolic markers in overweight/obese men. Clin Sci (Lond). 2016; 130:2073–82.
Article93. Yoshida H, Ishii M, Akagawa M. Propionate suppresses hepatic gluconeogenesis via GPR43/AMPK signaling pathway. Arch Biochem Biophys. 2019; 672:108057.
Article94. Al-Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R, et al. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest. 2012; 42:357–64.
Article95. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014; 156:84–96.
Article96. Chambers ES, Byrne CS, Aspey K, Chen Y, Khan S, Morrison DJ, et al. Acute oral sodium propionate supplementation raises resting energy expenditure and lipid oxidation in fasted humans. Diabetes Obes Metab. 2018; 20:1034–9.
Article97. Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009; 58:1509–17.
Article98. Mollica MP, Mattace Raso G, Cavaliere G, Trinchese G, De Filippo C, Aceto S, et al. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes. 2017; 66:1405–18.
Article99. Ohira H, Fujioka Y, Katagiri C, Mamoto R, Aoyama-Ishikawa M, Amako K, et al. Butyrate attenuates inflammation and lipolysis generated by the interaction of adipocytes and macrophages. J Atheroscler Thromb. 2013; 20:425–42.
Article100. Li Z, Yi CX, Katiraei S, Kooijman S, Zhou E, Chung CK, et al. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut. 2018; 67:1269–79.
Article101. De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Backhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 2016; 24:151–7.
Article102. Zhang L, Yang G, Untereiner A, Ju Y, Wu L, Wang R. Hydrogen sulfide impairs glucose utilization and increases gluconeogenesis in hepatocytes. Endocrinology. 2013; 154:114–26.
Article103. Virtue AT, McCright SJ, Wright JM, Jimenez MT, Mowel WK, Kotzin JJ, et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med. 2019; 11:eaav1892.
Article104. Hoyles L, Fernandez-Real JM, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018; 24:1070–80.
Article105. Goto T, Kim YI, Furuzono T, Takahashi N, Yamakuni K, Yang HE, et al. 10-Oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARγ and stimulates adipogenesis. Biochem Biophys Res Commun. 2015; 459:597–603.
Article106. Kim M, Furuzono T, Yamakuni K, Li Y, Kim YI, Takahashi H, et al. 10-Oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, enhances energy metabolism by activation of TRPV1. FASEB J. 2017; 31:5036–48.107. Takahashi Y, Kushiro M, Shinohara K, Ide T. Dietary conjugated linoleic acid reduces body fat mass and affects gene expression of proteins regulating energy metabolism in mice. Comp Biochem Physiol B Biochem Mol Biol. 2002; 133:395–404.
Article108. Park Y, Park Y. Conjugated fatty acids increase energy expenditure in part by increasing voluntary movement in mice. Food Chem. 2012; 133:400–9.
Article109. Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, et al. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006; 1761:736–44.
Article110. Houghton MJ, Kerimi A, Mouly V, Tumova S, Williamson G. Gut microbiome catabolites as novel modulators of muscle cell glucose metabolism. FASEB J. 2019; 33:1887–98.
Article111. Takagaki A, Yoshioka Y, Yamashita Y, Nagano T, Ikeda M, Hara-Terawaki A, et al. Effects of microbial metabolites of (-)-epigallocatechin gallate on glucose uptake in L6 skeletal muscle cell and glucose tolerance in ICR mice. Biol Pharm Bull. 2019; 42:212–21.
Article112. Choi Y, Kwon Y, Kim DK, Jeon J, Jang SC, Wang T, et al. Gut microbe-derived extracellular vesicles induce insulin resistance, thereby impairing glucose metabolism in skeletal muscle. Sci Rep. 2015; 5:15878.
Article113. Chen S, Henderson A, Petriello MC, Romano KA, Gearing M, Miao J, et al. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell Metab. 2019; 30:1141–51.
Article114. Gao X, Liu X, Xu J, Xue C, Xue Y, Wang Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J Biosci Bioeng. 2014; 118:476–81.
Article115. Aguirre M, Eck A, Koenen ME, Savelkoul PH, Budding AE, Venema K. Diet drives quick changes in the metabolic activity and composition of human gut microbiota in a validated in vitro gut model. Res Microbiol. 2016; 167:114–25.116. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016; 535:376–81.
Article117. Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018; 175:947–61.
Article118. Yoon HS, Cho CH, Yun MS, Jang SJ, You HJ, Kim JH, et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat Microbiol. 2021; 6:563–73.
Article119. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017; 23:107–13.
Article120. Martin AM, Yabut JM, Choo JM, Page AJ, Sun EW, Jessup CF, et al. The gut microbiome regulates host glucose homeostasis via peripheral serotonin. Proc Natl Acad Sci U S A. 2019; 116:19802–4.
Article121. Li X, Zhang B, Hu Y, Zhao Y. New insights into gut-bacteria-derived indole and its derivatives in intestinal and liver diseases. Front Pharmacol. 2021; 12:769501.
Article122. Aw W, Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. J Diabetes Investig. 2018; 9:5–12.
Article123. Mardinoglu A, Boren J, Smith U. Confounding effects of metformin on the human gut microbiome in type 2 diabetes. Cell Metab. 2016; 23:10–2.
Article124. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015; 528:262–6.
Article125. Devkota S. Microbiome. Prescription drugs obscure microbiome analyses. Science. 2016; 351:452–3.126. Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020; 11:362.
Article127. Kim DH. Gut microbiota-mediated drug-antibiotic interactions. Drug Metab Dispos. 2015; 43:1581–9.
Article128. Bryrup T, Thomsen CW, Kern T, Allin KH, Brandslund I, Jorgensen NR, et al. Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia. 2019; 62:1024–35.
Article129. Elbere I, Kalnina I, Silamikelis I, Konrade I, Zaharenko L, Sekace K, et al. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS One. 2018; 13:e0204317.
Article130. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017; 23:850–8.
Article131. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63:727–35.
Article132. Inzucchi SE, Bergenstal RM, Buse JB, Diamant M, Ferrannini E, Nauck M, et al. Management of hyperglycaemia in type 2 diabetes: a patient-centered approach. Position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia. 2012; 55:1577–96.
Article133. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017; 40:54–62.
Article134. Rosario D, Benfeitas R, Bidkhori G, Zhang C, Uhlen M, Shoaie S, et al. Understanding the representative gut microbiota dysbiosis in metformin-treated type 2 diabetes patients using genome-scale metabolic modeling. Front Physiol. 2018; 9:775.
Article135. Bai J, Zhu Y, Dong Y. Response of gut microbiota and inflammatory status to bitter melon (Momordica charantia L.) in high fat diet induced obese rats. J Ethnopharmacol. 2016; 194:717–26.
Article136. Xu P, Hong F, Wang J, Wang J, Zhao X, Wang S, et al. DBZ is a putative PPARγ agonist that prevents high fat diet-induced obesity, insulin resistance and gut dysbiosis. Biochim Biophys Acta Gen Subj. 2017; 1861:2690–701.
Article137. Stein SA, Lamos EM, Davis SN. A review of the efficacy and safety of oral antidiabetic drugs. Expert Opin Drug Saf. 2013; 12:153–75.
Article138. Lee DM, Battson ML, Jarrell DK, Hou S, Ecton KE, Weir TL, et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol. 2018; 17:62.
Article139. Olivares M, Neyrinck AM, Potgens SA, Beaumont M, Salazar N, Cani PD, et al. The DPP-4 inhibitor vildagliptin impacts the gut microbiota and prevents disruption of intestinal homeostasis induced by a Western diet in mice. Diabetologia. 2018; 61:1838–48.
Article140. Yan X, Feng B, Li P, Tang Z, Wang L. Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: an animal study. J Diabetes Res. 2016; 2016:2093171.
Article141. Zhang F, Wang M, Yang J, Xu Q, Liang C, Chen B, et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine. 2019; 66:485–93.
Article142. Su B, Liu H, Li J, Sunli Y, Liu B, Liu D, et al. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J Diabetes. 2015; 7:729–39.143. Zhang X, Fang Z, Zhang C, Xia H, Jie Z, Han X, et al. Effects of acarbose on the gut microbiota of prediabetic patients: a randomized, double-blind, controlled crossover trial. Diabetes Ther. 2017; 8:293–307.
Article144. Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021; 27:321–32.
Article145. Beghini F, Pasolli E, Truong TD, Putignani L, Caccio SM, Segata N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017; 11:2848–63.146. Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016; 540:544–51.
Article147. Leshem A, Segal E, Elinav E. The gut microbiome and individual-specific responses to diet. mSystems. 2020; 5:e00665–20.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Systems Biology: A Multi-Omics Integration Approach to Metabolism and the Microbiome
- Complex influences of gut microbiome metabolism on various drug responses
- Human microbiome studies in Korea
- Emerging Relationship between the Gut Microbiome and Prostate Cancer
- The Urinary Microbiome: A Pediatric Urological Perspective