J Korean Med Sci.
2024 Aug;39(31):e223. 10.3346/jkms.2024.39.e223.
Systemic Inflammatory Proteomic Biomarkers in Atopic Dermatitis: Exploring Potential Indicators for Disease Severity
- Affiliations
-
- 1Department of Dermatology, Incheon St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2Samsung Genome Institute, Samsung Medical Center, Seoul, Korea
- 3Department of Biomedical Sciences, Seoul National University Graduate School, Seoul, Korea
- 4Department of Dermatology, Eunpyeong St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 5Department of Dermatology, Seoul National University Hospital, Seoul, Korea
- KMID: 2558524
- DOI: http://doi.org/10.3346/jkms.2024.39.e223
Abstract
- Background
Atopic dermatitis (AD) is a chronic inflammatory cutaneous disorder, that emerges from intricate interplays among genetic predisposition, immune dysregulation, environmental factors, and compromised skin barrier. Understanding the inflammatory pathway in AD is important due to its fundamental role in the pathogenesis of AD. This study aimed to explore the diverse spectrum of proteins linked to the inflammation of AD and the relationship between systemic biomarkers and clinical severity in AD.
Methods
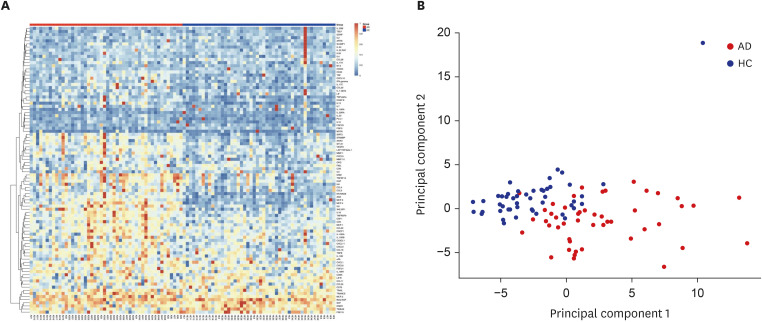
We examined the blood samples from 48 patients with AD and 48 healthy controls (HCs) using the Proximity Extension Assay (Olink). Differentially expressed proteins (DEPs) were identified and Pearson correlation analysis was conducted to determine systemic proteomic biomarkers associated with severity of AD.
Results
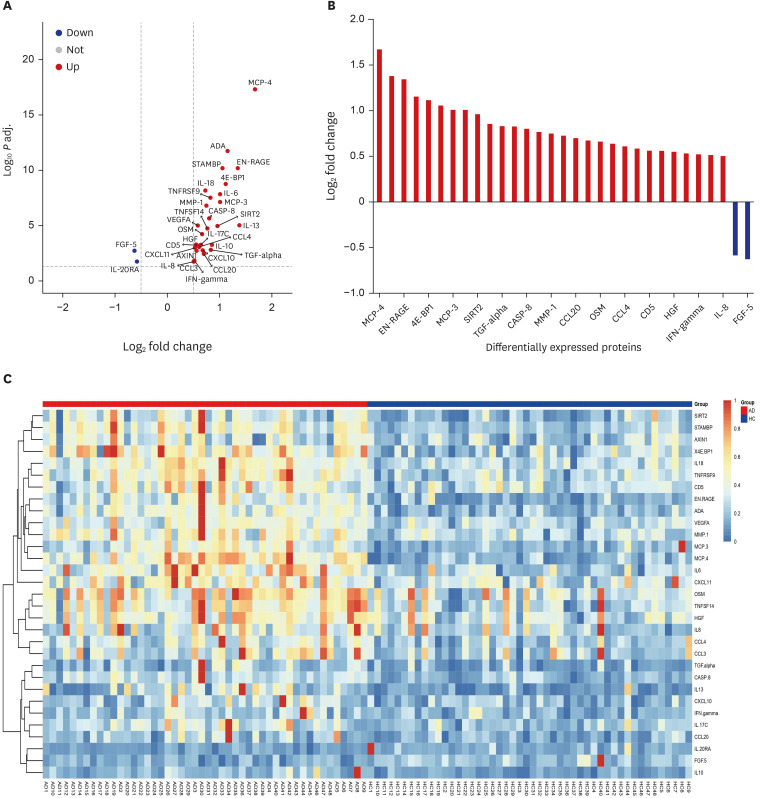
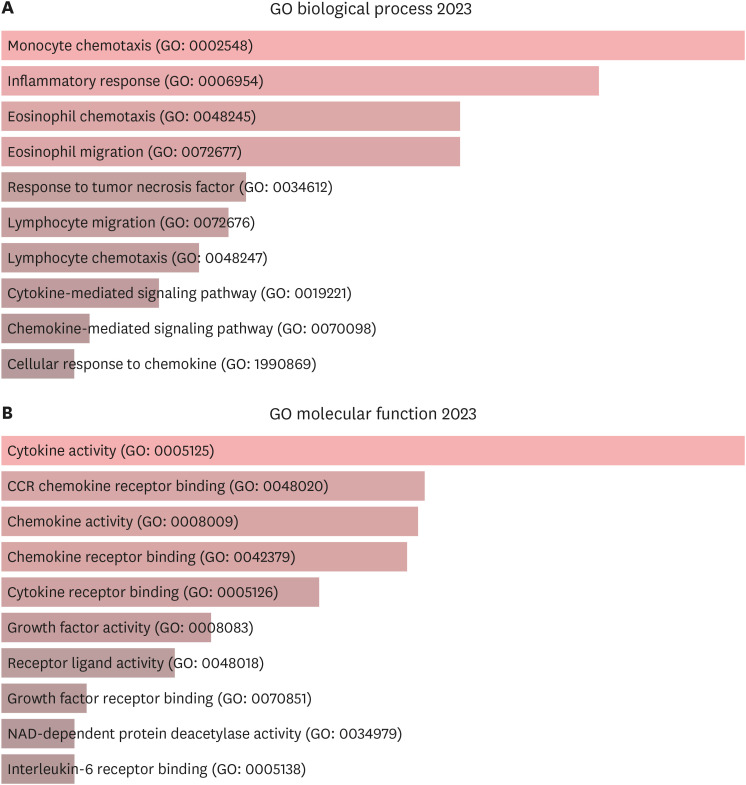
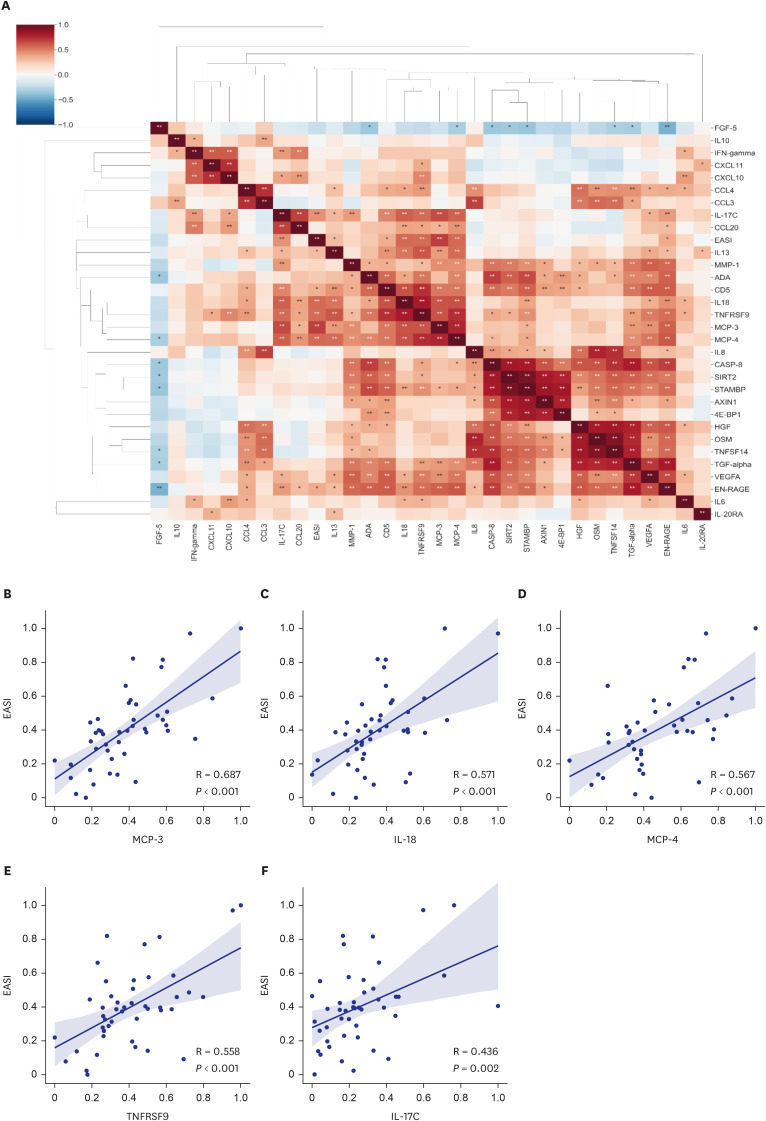
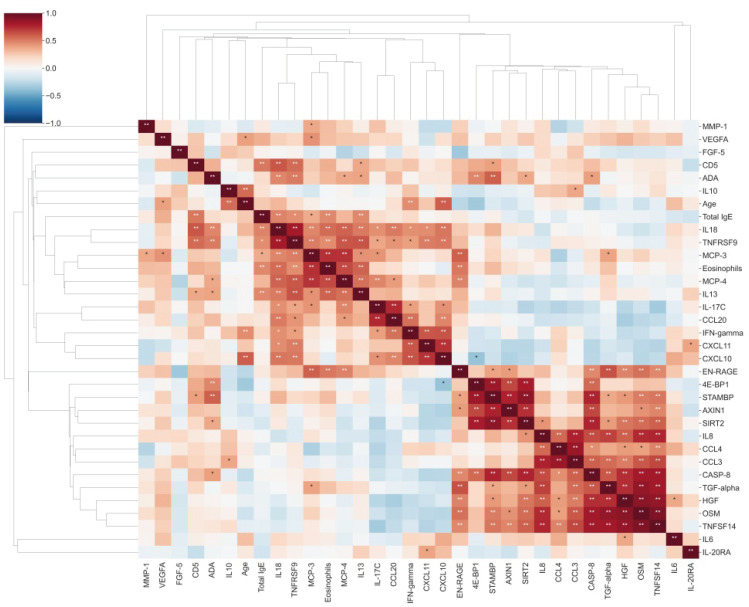
A total of 29 DEPs were significantly up-regulated and 2 DEPs were significantly down-regulated in AD compared with the HC. The MCP-4, IL-18, MCP-3, TNFRSF9, and IL-17C were the top 5 highest DEPs associated with the severity of AD.
Conclusion
Our study sheds light on the intricate network of inflammatory proteins in AD and their potential implications for disease severity. Our results indicate that these systemic inflammatory proteins could be valuable for assessing AD severity and enhancing our understanding of the disease's complexity and its potential management strategies.
Keyword
Figure
Reference
-
1. Leung DY. Pathogenesis of atopic dermatitis. J Allergy Clin Immunol. 1999; 104(3 Pt 2):S99–108. PMID: 10482860.2. Dubin C, Del Duca E, Guttman-Yassky E. The IL-4, IL-13 and IL-31 pathways in atopic dermatitis. Expert Rev Clin Immunol. 2021; 17(8):835–852. PMID: 34106037.3. Akdis CA, Arkwright PD, Brüggen MC, Busse W, Gadina M, Guttman-Yassky E, et al. Type 2 immunity in the skin and lungs. Allergy. 2020; 75(7):1582–1605. PMID: 32319104.4. Renert-Yuval Y, Thyssen JP, Bissonnette R, Bieber T, Kabashima K, Hijnen D, et al. Biomarkers in atopic dermatitis-a review on behalf of the International Eczema Council. J Allergy Clin Immunol. 2021; 147(4):1174–1190.e1. PMID: 33516871.5. Sanyal RD, Pavel AB, Glickman J, Chan TC, Zheng X, Zhang N, et al. Atopic dermatitis in African American patients is TH2/TH22-skewed with TH1/TH17 attenuation. Ann Allergy Asthma Immunol. 2019; 122(1):99–110.e6. PMID: 30223113.6. Kaufman BP, Guttman-Yassky E, Alexis AF. Atopic dermatitis in diverse racial and ethnic groups-variations in epidemiology, genetics, clinical presentation and treatment. Exp Dermatol. 2018; 27(4):340–357. PMID: 29457272.7. Noda S, Suárez-Fariñas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. 2015; 136(5):1254–1264. PMID: 26428954.8. Czarnowicki T, He H, Krueger JG, Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. 2019; 143(1):1–11. PMID: 30612663.9. Fredman G, Skov L, Mann M, Dyring-Andersen B. Towards precision dermatology: emerging role of proteomic analysis of the skin. Dermatology. 2022; 238(2):185–194. PMID: 34062531.10. Pavel AB, Zhou L, Diaz A, Ungar B, Dan J, He H, et al. The proteomic skin profile of moderate-to-severe atopic dermatitis patients shows an inflammatory signature. J Am Acad Dermatol. 2020; 82(3):690–699. PMID: 31669080.11. Park YL, Kim HD, Kim KH, Kim MN, Kim JW, Ro YS, et al. Report from ADRG: a study on the diagnostic criteria of Korean atopic dermatitis. Korean J Dermatol. 2006; 44(6):659–663.12. Kim JE, Shin MK, Park GH, Lee UH, Lee JH, Han TY, et al. 2019 Consensus Korean diagnostic guidelines to define severity classification and treatment refractoriness for atopic dermatitis: objective and subjective assessment of severity. Ann Dermatol. 2019; 31(6):654–661. PMID: 33911665.13. R Foundation for Statistical Computing. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing;2023.14. Xie Z, Bailey A, Kuleshov MV, Clarke DJ, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021; 1(3):e90. PMID: 33780170.15. Sims JT, Chang CY, Higgs RE, Engle SM, Liu Y, Sissons SE, et al. Insights into adult atopic dermatitis heterogeneity derived from circulating biomarker profiling in patients with moderate-to-severe disease. Exp Dermatol. 2021; 30(11):1650–1661. PMID: 34003519.16. He H, Del Duca E, Diaz A, Kim HJ, Gay-Mimbrera J, Zhang N, et al. Mild atopic dermatitis lacks systemic inflammation and shows reduced nonlesional skin abnormalities. J Allergy Clin Immunol. 2021; 147(4):1369–1380. PMID: 33011244.17. Ungar B, Garcet S, Gonzalez J, Dhingra N, Correa da Rosa J, Shemer A, et al. An integrated model of atopic dermatitis biomarkers highlights the systemic nature of the disease. J Invest Dermatol. 2017; 137(3):603–613. PMID: 27825969.18. Mendez-Enriquez E, García-Zepeda EA. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology. 2013; 21(6):397–406. PMID: 23846739.19. Ihim SA, Abubakar SD, Zian Z, Sasaki T, Saffarioun M, Maleknia S, et al. Interleukin-18 cytokine in immunity, inflammation, and autoimmunity: biological role in induction, regulation, and treatment. Front Immunol. 2022; 13:919973. PMID: 36032110.20. Lee JH, Cho DH, Park HJ. IL-18 and cutaneous inflammatory diseases. Int J Mol Sci. 2015; 16(12):29357–29369. PMID: 26690141.21. Yang G, Seok JK, Kang HC, Cho YY, Lee HS, Lee JY. Skin barrier abnormalities and immune dysfunction in atopic dermatitis. Int J Mol Sci. 2020; 21(8):2867. PMID: 32326002.22. Lyubchenko T, Collins HK, Goleva E, Leung DY. Skin tape sampling technique identifies proinflammatory cytokines in atopic dermatitis skin. Ann Allergy Asthma Immunol. 2021; 126(1):46–53.e2. PMID: 32896640.23. Hoshino T, Wiltrout RH, Young HA. IL-18 is a potent coinducer of IL-13 in NK and T cells: a new potential role for IL-18 in modulating the immune response. J Immunol. 1999; 162(9):5070–5077. PMID: 10227975.24. Hayashi N, Yoshimoto T, Izuhara K, Matsui K, Tanaka T, Nakanishi K. T helper 1 cells stimulated with ovalbumin and IL-18 induce airway hyperresponsiveness and lung fibrosis by IFN-γ and IL-13 production. Proc Natl Acad Sci U S A. 2007; 104(37):14765–14770. PMID: 17766435.25. Shi HZ, Qin XJ. CD4CD25 regulatory T lymphocytes in allergy and asthma. Allergy. 2005; 60(8):986–995. PMID: 15969678.26. Wong HY, Schwarz H. CD137/CD137 ligand signalling regulates the immune balance: a potential target for novel immunotherapy of autoimmune diseases. J Autoimmun. 2020; 112:102499. PMID: 32505443.27. Ma L, Xue HB, Guan XH, Shu CM, Zhang JH, Yu J. Possible pathogenic role of T helper type 9 cells and interleukin (IL)-9 in atopic dermatitis. Clin Exp Immunol. 2014; 175(1):25–31. PMID: 24032555.28. Guttman-Yassky E, Krueger JG. IL-17C: a unique epithelial cytokine with potential for targeting across the spectrum of atopic dermatitis and psoriasis. J Invest Dermatol. 2018; 138(7):1467–1469. PMID: 29941097.29. Wang S, Zhu R, Gu C, Zou Y, Yin H, Xu J, et al. Distinct clinical features and serum cytokine pattern of elderly atopic dermatitis in China. J Eur Acad Dermatol Venereol. 2020; 34(10):2346–2352. PMID: 32163633.30. He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, et al. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol. 2021; 147(1):199–212. PMID: 32709423.31. Vandeghinste N, Klattig J, Jagerschmidt C, Lavazais S, Marsais F, Haas JD, et al. Neutralization of IL-17C reduces skin inflammation in mouse models of psoriasis and atopic dermatitis. J Invest Dermatol. 2018; 138(7):1555–1563. PMID: 29474945.32. Simon D, Braathen LR, Simon HU. Eosinophils and atopic dermatitis. Allergy. 2004; 59(6):561–570. PMID: 15147438.33. Garcia-Zepeda EA, Combadiere C, Rothenberg ME, Sarafi MN, Lavigne F, Hamid Q, et al. Human monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine with activities on monocytes, eosinophils, and basophils induced in allergic and nonallergic inflammation that signals through the CC chemokine receptors (CCR)-2 and -3. J Immunol. 1996; 157(12):5613–5626. PMID: 8955214.34. Shang XZ, Chiu BC, Stolberg V, Lukacs NW, Kunkel SL, Murphy HS, et al. Eosinophil recruitment in type-2 hypersensitivity pulmonary granulomas: source and contribution of monocyte chemotactic protein-3 (CCL7). Am J Pathol. 2002; 161(1):257–266. PMID: 12107110.35. Kandikattu HK, Upparahalli Venkateshaiah S, Mishra A. Synergy of interleukin (IL)-5 and IL-18 in eosinophil mediated pathogenesis of allergic diseases. Cytokine Growth Factor Rev. 2019; 47:83–98. PMID: 31126874.36. Brandt EB, Lewkowich IP. RAGE-induced asthma: a role for the receptor for advanced glycation end-products in promoting allergic airway disease. J Allergy Clin Immunol. 2019; 144(3):651–653. PMID: 31251951.37. Perkins TN, Donnell ML, Oury TD. The axis of the receptor for advanced glycation endproducts in asthma and allergic airway disease. Allergy. 2021; 76(5):1350–1366. PMID: 32976640.38. Bikah G, Lynd FM, Aruffo AA, Ledbetter JA, Bondada S. A role for CD5 in cognate interactions between T cells and B cells, and identification of a novel ligand for CD5. Int Immunol. 1998; 10(8):1185–1196. PMID: 9723705.39. de Vries JE, Zurawski G. Immunoregulatory properties of IL-13: its potential role in atopic disease. Int Arch Allergy Immunol. 1995; 106(3):175–179. PMID: 7888780.40. Yoshimoto T, Mizutani H, Tsutsui H, Noben-Trauth N, Yamanaka K, Tanaka M, et al. IL-18 induction of IgE: dependence on CD4+ T cells, IL-4 and STAT6. Nat Immunol. 2000; 1(2):132–137. PMID: 11248805.41. Wirtz MK, Keller KE. The role of the IL-20 subfamily in glaucoma. Mediators Inflamm. 2016; 2016:4083735. PMID: 26903709.42. Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nat Rev Immunol. 2014; 14(12):783–795. PMID: 25421700.43. Kutwin M, Woźniacka A.. Interleukins 20 and 8 - less widely known cytokines in psoriasis. Postepy Dermatol Alergol. 2023; 40(2):194–203. PMID: 37312911.44. Lu Z, Xiao S, Chen W, Zhu R, Yang H, Steinhoff M, et al. IL-20 promotes cutaneous inflammation and peripheral itch sensation in atopic dermatitis. FASEB J. 2022; 36(6):e22334. PMID: 35486004.45. Werner S, Weinberg W, Liao X, Peters KG, Blessing M, Yuspa SH, et al. Targeted expression of a dominant-negative FGF receptor mutant in the epidermis of transgenic mice reveals a role of FGF in keratinocyte organization and differentiation. EMBO J. 1993; 12(7):2635–2643. PMID: 7687538.46. Yang J, Meyer M, Müller AK, Böhm F, Grose R, Dauwalder T, et al. Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J Cell Biol. 2010; 188(6):935–952. PMID: 20308431.47. Engle SM, Chang CY, Ulrich BJ, Satterwhite A, Hayes T, Robling K, et al. Predictive biomarker modeling of pediatric atopic dermatitis severity based on longitudinal serum collection. Clin Exp Immunol. 2022; 207(3):253–262.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effect of Antigen Sensitization and Development of Respiratory Allergy Disease on Severity of Atopic Dermatitis
- The KAPARD guidelines for atopic dermatitis in children and adolescents: Part II. Systemic treatment, novel therapeutics, and adjuvant therapy
- Measurement of Atopic Dermatitis Disability
- Improvement of Post-inflammatory Hyperpigmentation, Subsequent to Cold Atmospheric Plasma Treatment, in a Patient with Atopic Dermatitis
- Childhood Systemic Lupus Erythematosus Misdiagnosed as Atopic Dermatitis