J Korean Med Sci.
2024 Jul;39(28):e206. 10.3346/jkms.2024.39.e206.
Genotype Analysis of Respiratory Syncytial Virus Before and After the COVID-19 Pandemic Using WholeGenome Sequencing: A Prospective, Single-Center Study in Korea From 2019 to 2022
- Affiliations
-
- 1Department of Pediatrics, Severance Children’s Hospital, Yonsei University College of Medicine, Seoul, Korea
- 2Institute for Immunology and Immunological Diseases, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea
- 4Dxome Co., Ltd., Seongnam, Korea
- KMID: 2558244
- DOI: http://doi.org/10.3346/jkms.2024.39.e206
Abstract
- Background
Respiratory syncytial virus (RSV), a highly transmissible virus, is the leading cause of lower respiratory tract infections. We examined molecular changes in the RSV genome before and after the coronavirus disease 2019 (COVID-19) pandemic in Korea, and investigated whether drug-resistant mutations were present.
Methods
In this prospective, single-center study, RSV-positive respiratory samples were collected between September 2019 and December 2022. Long-read whole-genome sequencing (WGS) was performed, and the presence of known drug-resistant substitutions for palivizumab, nirsevimab, and suptavumab was investigated.
Results
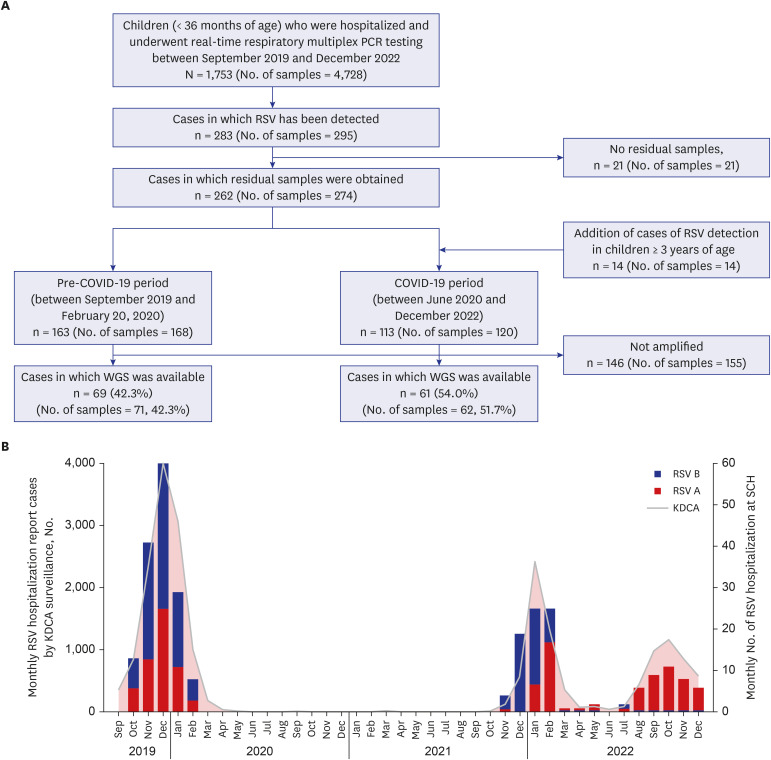
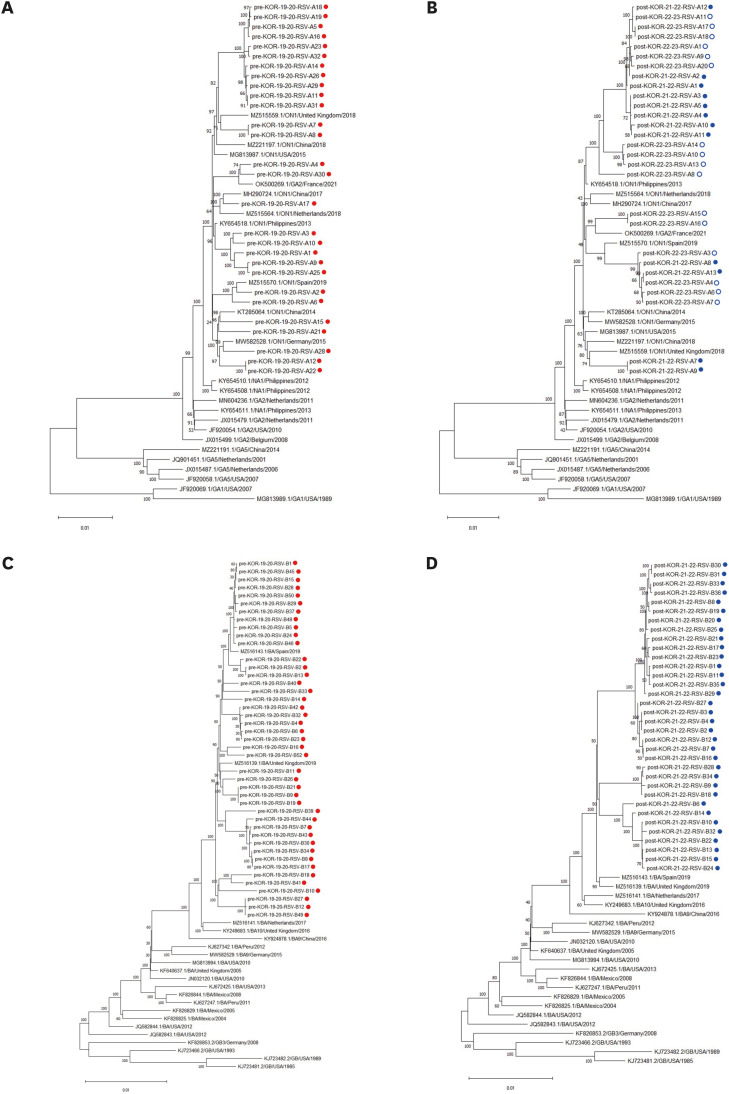
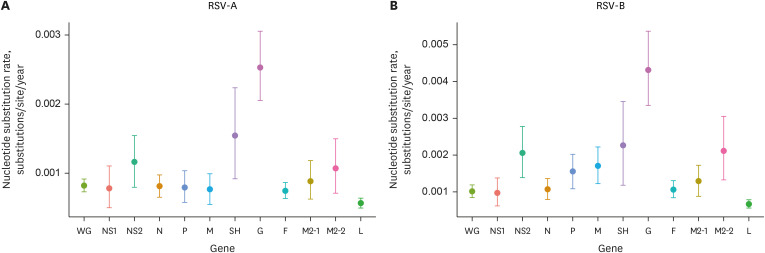
Overall, 288 respiratory samples were collected from 276 children. WGS data were available for 133 samples (71 and 62 samples from the pre- and post-pandemic periods, respectively). All RSV-A strains (n = 56) belonged to the GA2.3.5 (ON1) genotype, whereas all RSV-B strains (n = 77) belonged to the GB5.0.5a (BA) genotype. No significant differences in genotypes were observed between the pre- and post-pandemic periods. In addition, no notable mutations related to nirsevimab or palivizumab resistance were detected in the F gene. However, the L172Q and S173L substitutions, which are known to confer resistance to suptavumab, were present in all RSV-B samples.
Conclusion
Despite the unprecedented interruption of RSV seasonality, there were no significant molecular changes in circulating RSV strains in Korea related to nirsevimab or palivizumab resistance before and after the COVID-19 pandemic. However, RSV-specific drug-resistance substitutions for suptavumab were identified.
Keyword
Figure
Reference
-
1. Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009; 360(6):588–598. PMID: 19196675.2. Li Y, Wang X, Blau DM, Caballero MT, Feikin DR, Gill CJ, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. 2022; 399(10340):2047–2064. PMID: 35598608.3. Savic M, Penders Y, Shi T, Branche A, Pirçon JY. Respiratory syncytial virus disease burden in adults aged 60 years and older in high-income countries: a systematic literature review and meta-analysis. Influenza Other Respi Viruses. 2023; 17(1):e13031.4. Mejias A, Garcia-Maurino C, Rodriguez-Fernandez R, Peeples ME, Ramilo O. Development and clinical applications of novel antibodies for prevention and treatment of respiratory syncytial virus infection. Vaccine. 2017; 35(3):496–502. PMID: 27692523.5. Tabor DE, Fernandes F, Langedijk AC, Wilkins D, Lebbink RJ, Tovchigrechko A, et al. Global molecular epidemiology of respiratory syncytial virus from the 2017-2018 INFORM-RSV study. J Clin Microbiol. 2020; 59(1):e01828-20. PMID: 33087438.6. Zhu Q, Patel NK, McAuliffe JM, Zhu W, Wachter L, McCarthy MP, et al. Natural polymorphisms and resistance-associated mutations in the fusion protein of respiratory syncytial virus (RSV): effects on RSV susceptibility to palivizumab. J Infect Dis. 2012; 205(4):635–638. PMID: 22184728.7. Yu JM, Fu YH, Peng XL, Zheng YP, He JS. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci Rep. 2021; 11(1):12941. PMID: 34155268.8. Ramilo O, Rodriguez-Fernandez R, Mejias A. Respiratory syncytial virus infection: old challenges and new approaches. J Infect Dis. 2023; 228(1):4–7. PMID: 36715631.9. Teirlinck AC, Johannesen CK, Broberg EK, Penttinen P, Campbell H, Nair H, et al. New perspectives on respiratory syncytial virus surveillance at the national level: lessons from the COVID-19 pandemic. Eur Respir J. 2023; 61(4):2201569. PMID: 37012081.10. Huh K, Jung J, Hong J, Kim M, Ahn JG, Kim JH, et al. Impact of nonpharmaceutical interventions on the incidence of respiratory infections during the coronavirus disease 2019 (COVID-19) outbreak in Korea: a nationwide surveillance study. Clin Infect Dis. 2021; 72(7):e184–e191. PMID: 33150393.11. Kim JH, Roh YH, Ahn JG, Kim MY, Huh K, Jung J, et al. Respiratory syncytial virus and influenza epidemics disappearance in Korea during the 2020-2021 season of COVID-19. Int J Infect Dis. 2021; 110:29–35. PMID: 34245886.12. Kim JH, Kim HY, Lee M, Ahn JG, Baek JY, Kim MY, et al. Respiratory syncytial virus outbreak without influenza in the second year of the coronavirus disease 2019 pandemic: a national sentinel surveillance in Korea, 2021-2022 season. J Korean Med Sci. 2022; 37(34):e258. PMID: 36038956.13. Choi SH, Park KS, Kim YJ. Analysis of respiratory syncytial virus fusion protein from clinical isolates of Korean children in palivizumab era, 2009-2015. J Infect Chemother. 2019; 25(7):514–519. PMID: 30885457.14. Wilkins D, Langedijk AC, Lebbink RJ, Morehouse C, Abram ME, Ahani B, et al. Nirsevimab binding-site conservation in respiratory syncytial virus fusion glycoprotein worldwide between 1956 and 2021: an analysis of observational study sequencing data. Lancet Infect Dis. 2023; 23(7):856–866. PMID: 36940703.15. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012; 29(8):1969–1973. PMID: 22367748.16. Ahani B, Tuffy KM, Aksyuk AA, Wilkins D, Abram ME, Dagan R, et al. Molecular and phenotypic characteristics of RSV infections in infants during two nirsevimab randomized clinical trials. Nat Commun. 2023; 14(1):4347. PMID: 37468530.17. Yasui Y, Yamaji Y, Sawada A, Ito T, Nakayama T. Cell fusion assay by expression of respiratory syncytial virus (RSV) fusion protein to analyze the mutation of palivizumab-resistant strains. J Virol Methods. 2016; 231:48–55. PMID: 26794681.18. Yun KW, Choi EH, Lee HJ. Molecular epidemiology of respiratory syncytial virus for 28 consecutive seasons (1990-2018) and genetic variability of the duplication region in the G gene of genotypes ON1 and BA in South Korea. Arch Virol. 2020; 165(5):1069–1077. PMID: 32144544.19. Kim HN, Hwang J, Yoon SY, Lim CS, Cho Y, Lee CK, et al. Molecular characterization of human respiratory syncytial virus in Seoul, South Korea, during 10 consecutive years, 2010-2019. PLoS One. 2023; 18(4):e0283873. PMID: 37023101.20. Eden JS, Sikazwe C, Xie R, Deng YM, Sullivan SG, Michie A, et al. Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat Commun. 2022; 13(1):2884. PMID: 35610217.21. Lin TY, Chi H, Kuo CY, Tsai HP, Wang JR, Liu CC, et al. Outbreak of respiratory syncytial virus subtype ON1 among children during COVID-19 pandemic in Southern Taiwan. J Microbiol Immunol Infect. 2022; 55(6 Pt 2):1168–1179. PMID: 36137926.22. Pierangeli A, Nenna R, Fracella M, Scagnolari C, Oliveto G, Sorrentino L, et al. Genetic diversity and its impact on disease severity in respiratory syncytial virus subtype-A and -B bronchiolitis before and after pandemic restrictions in Rome. J Infect. 2023; 87(4):305–314. PMID: 37495189.23. Simões EA, Forleo-Neto E, Geba GP, Kamal M, Yang F, Cicirello H, et al. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin Infect Dis. 2021; 73(11):e4400–e4408. PMID: 32897368.24. Li W, Wang Y, Yu B, Tan Q, Zhou J, Hu J, et al. Disease severity of respiratory syncytial virus (RSV) infection correlate to a novel set of five amino acid substitutions in the RSV attachment glycoprotein (G) in China. Virus Res. 2020; 281:197937. PMID: 32194139.25. Midulla F, Nenna R, Scagnolari C, Petrarca L, Frassanito A, Viscido A, et al. How respiratory syncytial virus genotypes influence the clinical course in infants hospitalized for bronchiolitis. J Infect Dis. 2019; 219(4):526–534. PMID: 30204889.26. Abu-Raya B, Viñeta Paramo M, Reicherz F, Lavoie PM. Why has the epidemiology of RSV changed during the COVID-19 pandemic? EClinicalMedicine. 2023; 61:102089. PMID: 37483545.27. Gueudin M, Vabret A, Petitjean J, Gouarin S, Brouard J, Freymuth F. Quantitation of respiratory syncytial virus RNA in nasal aspirates of children by real-time RT-PCR assay. J Virol Methods. 2003; 109(1):39–45. PMID: 12668266.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Shift in Clinical Epidemiology of Human Parainfluenza Virus Type 3 and Respiratory Syncytial Virus B Infections in Korean Children Before and During the COVID-19 Pandemic: A Multicenter Retrospective Study
- Reemergence of Parainfluenza Virus Type 3 and Respiratory Syncytial Virus Infections During the COVID-19 Pandemic
- COVID-19 and Fungal Infection
- Characteristics of Respiratory Syncytial Virus Infection in Hospitalized Children Before and During the COVID-19 Pandemic in Thailand
- Severe acute respiratory syndrome coronavirus 2 and respiratory syncytial virus coinfection in children