Cancer Res Treat.
2024 Jul;56(3):765-773. 10.4143/crt.2023.1294.
Analytical and Clinical Validation of a Highly Sensitive NGS-Based ctDNA Assay with Real-World Concordance in Non–Small Cell Lung Cancer
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Cancer Research Institute, Seoul National University, Seoul, Korea
- 3IMBdx, Seoul, Korea
- 4Department of Chemistry, Yonsei University, Seoul, Korea
- KMID: 2557663
- DOI: http://doi.org/10.4143/crt.2023.1294
Abstract
- Purpose
There have been needs to improve the sensitivity of liquid biopsy. This report aims to report the analytical and clinical validation of a next-generation sequencing (NGS)–based circulating tumor DNA (ctDNA) assay.
Materials and Methods
Analytical validation was conducted in vitro by evaluating the limit of detection (LOD), precision, and specificity for various genomic aberrations. The real-world performance in non–small cell lung cancer (NSCLC) was assessed by comparing the results of AlphaLiquid100 to the tissue-based results.
Results
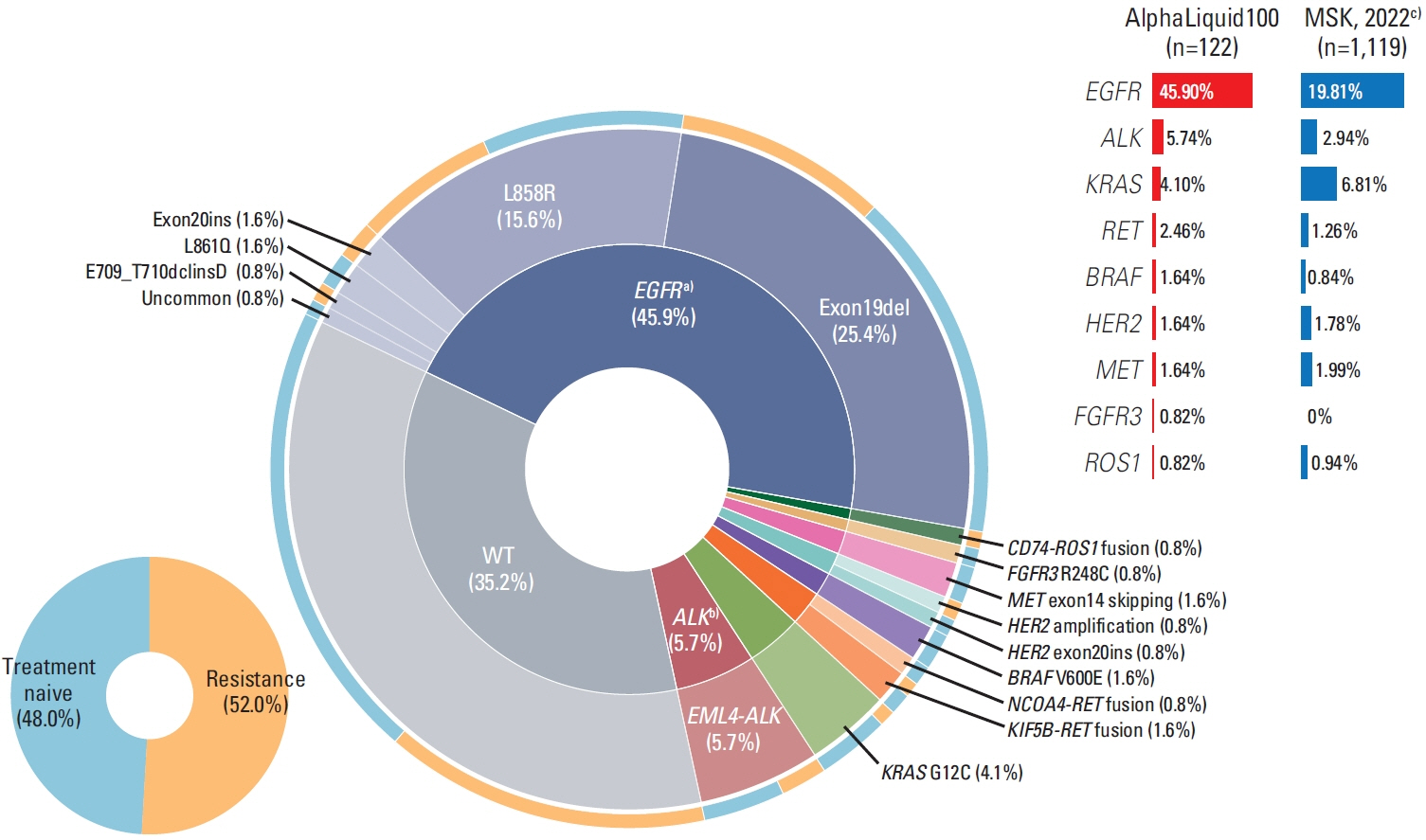
The LODs with 30 ng input DNA were 0.11%, 0.11%, 0.06%, 0.21%, and 2.13 copies for detecting single nucleotide variants, insertions, deletions, fusions, and copy number alterations (CNA), respectively. Quantitatively, single nucleotide variants/insertions and deletions, fusions, and CNAs showed a good correlation (R2=0.91, 0.40, and 0.65; y=0.95, 1.06, and 1.19) to the manufacturer’s values, and per-base specificities for all types of variants were near 100%. In real-world NSCLC (n=122), key actionable mutations in NSCLC were detected in 60.7% (74/122) with the ctDNA assay. Comparative analysis against the NGS-based tissue results for all key mutations showed positive percent agreement (PPA) of 85.3%. For individual genes, the PPA was as high as 95.7% for epidermal growth factor receptor (EGFR) mutations and 83.3% for ALK translocations. AlphaLiquid100 detected drug-sensitive EGFR mutation at a variant allele frequency as low as 0.02% and also identified an EGFR mutation in a case where tissue sample missed. Blood samples collected post-targeted therapies revealed additional acquired mutations.
Conclusion
The AlphaLiquid100 ctDNA assay demonstrates robust analytical validity, offering clinically important information for NSCLC patients.
Keyword
Figure
Reference
-
References
1. Alix-Panabieres C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021; 11:858–73.
Article2. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.
Article3. Thompson JC, Aggarwal C, Wong J, Nimgaonkar V, Hwang WT, Andronov M, et al. Plasma genotyping at the time of diagnostic tissue biopsy decreases time-to-treatment in patients with advanced NSCLC: results from a prospective pilot study. JTO Clin Res Rep. 2022; 3:100301.4. Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020; 26:1859–64.
Article5. National Comprehensive Cancer Network. Non-small cell lung cancer (version 3.2023) [Internet]. Plymouth Meeting, PA: National Comprehensive Cancer Network;2023 [cited 2023 Dec 5]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.6. Rolfo C, Mack P, Scagliotti GV, Aggarwal C, Arcila ME, Barlesi F, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the International Association for the Study of Lung Cancer. J Thorac Oncol. 2021; 16:1647–62.
Article7. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–60.8. Im SW, Chae J, Jang SS, Choi J, Yun J, Cha S, et al. A newly developed capture-based sequencing panel for genomic assay of lung cancer. Genes Genomics. 2020; 42:751–9.
Article9. Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023; 34:339–57.
Article10. Jee J, Lebow ES, Yeh R, Das JP, Namakydoust A, Paik PK, et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med. 2022; 28:2353–63.11. Song A, Kim TM, Kim DW, Kim S, Keam B, Lee SH, et al. Molecular changes associated with acquired resistance to crizotinib in ROS1-rearranged non-small cell lung cancer. Clin Cancer Res. 2015; 21:2379–87.12. De Mattos-Arruda L, Siravegna G. How to use liquid biopsies to treat patients with cancer. ESMO Open. 2021; 6:100060.
Article13. Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022; 21:79.
Article14. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for sdvanced non-dmall vell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018; 13:1248–68.15. U.S. Food and Drug Administration. List of cleared or approved companion diagnostic devices (in vitro and imaging tools) [Internet]. Silver Spring, MD: U.S. Food and Drug Administration; 2023 [cited 2023 Dec 5]. Available from: https://www.fda.gov/medical-devices/in-vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-in-vitro-and-imaging-tools.16. Sugimoto A, Matsumoto S, Udagawa H, Itotani R, Usui Y, Umemura S, et al. A large-scale prospective voncordance dtudy of plasma- and tissue-based next-generation targeted sequencing for advanced non-small cell lung cancer (LCSCRUM-Liquid). Clin Cancer Res. 2023; 29:1506–14.17. Desmeules P, Dusselier M, Bouffard C, Bafaro J, Fortin M, Labbe C, et al. Retrospective assessment of complementary liquid biopsy on tissue single-gene testing for tumor genotyping in advanced NSCLC. Curr Oncol. 2023; 30:575–85.
Article18. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020; 15:e0237802.
Article19. Yu C, Han Y, Wang M, Hua P, Zhang Y, Wang B. Concordance of ctDNA and tissue mutations in NSCLC: a meta-analysis. Cell Mol Biol (Noisy-le-grand). 2023; 69:89–95.
Article20. Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015; 21:3196–203.21. Jeong SH, Kyung D, Yuk HD, Jeong CW, Lee W, Yoon JK, et al. Practical utility of liquid biopsies for evaluating genomic alterations in castration-resistant prostate cancer. Cancers (Basel). 2023; 15:2847.
Article22. Kang JK, Heo S, Kim HP, Song SH, Yun H, Han SW, et al. Liquid biopsy-based tumor profiling for metastatic colorectal cancer patients with ultra-deep targeted sequencing. PLoS One. 2020; 15:e0232754.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Circulating Tumor DNA Testing for Precision Oncology
- Tissue and Plasma-Based Highly Sensitive Blocker Displacement Amplicon Nanopore Sequencing for EGFR Mutations in Lung Cancer
- Robust home brew fragment sizing assay for detection of MET exon 14 skipping mutation in non–small cell lung cancer patients in resource constrained community hospitals
- Chemotherapy for Small Cell Lung Cancer
- Utilizing Plasma Circulating Tumor DNA Sequencing for Precision Medicine in the Management of Solid Cancers