J Pathol Transl Med.
2021 Sep;55(5):324-329. 10.4132/jptm.2021.07.15.
Robust home brew fragment sizing assay for detection of MET exon 14 skipping mutation in non–small cell lung cancer patients in resource constrained community hospitals
- Affiliations
-
- 1Department of Laboratory Services, Molecular Diagnostics and Transfusion Medicine, Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India
- 2Section of Molecular Diagnostics, Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India
- KMID: 2520171
- DOI: http://doi.org/10.4132/jptm.2021.07.15
Abstract
- Background
A mutation/deletion involving donor or acceptor sites for exon 14 results in splicing out of exon 14 of the mesenchymal epithelial transition (MET) gene and is known as “MET exon 14 skipping” (ΔMET14). The two recent approvals with substantial objective responses and improved progression-free survival to MET inhibitors namely capmatinib and tepotinib necessitate the identification of this alteration upfront. We herein describe our experience of ΔMET14 detection by an mRNA-based assay using polymerase chain reaction followed by fragment sizing.
Methods
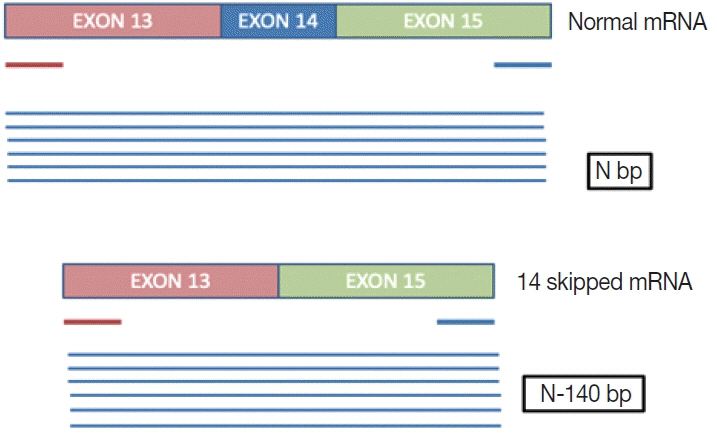
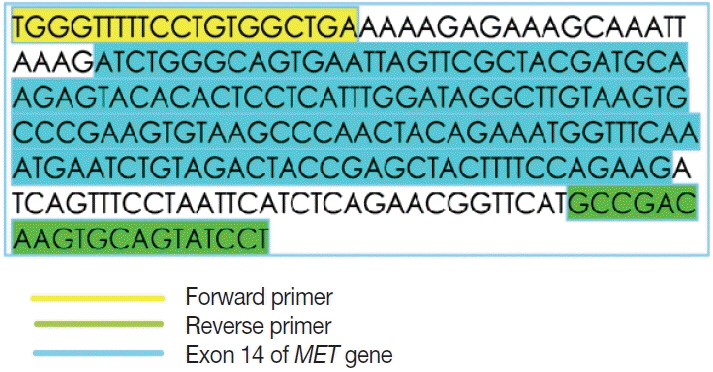
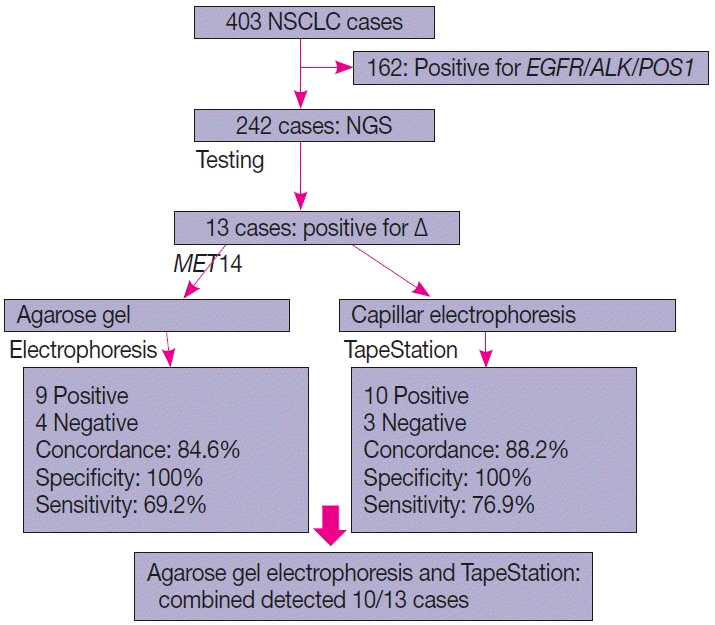
This is a home brew assay which was developed with the concept that the transcripts from true ΔMET14 will be shorter by ~140 bases than their wild type counterparts. The cases which were called MET exon 14 skipping positive on next-generation sequencing (NGS) were subjected to this assay, along with 13 healthy controls in order to establish the validity for true negatives.
Results
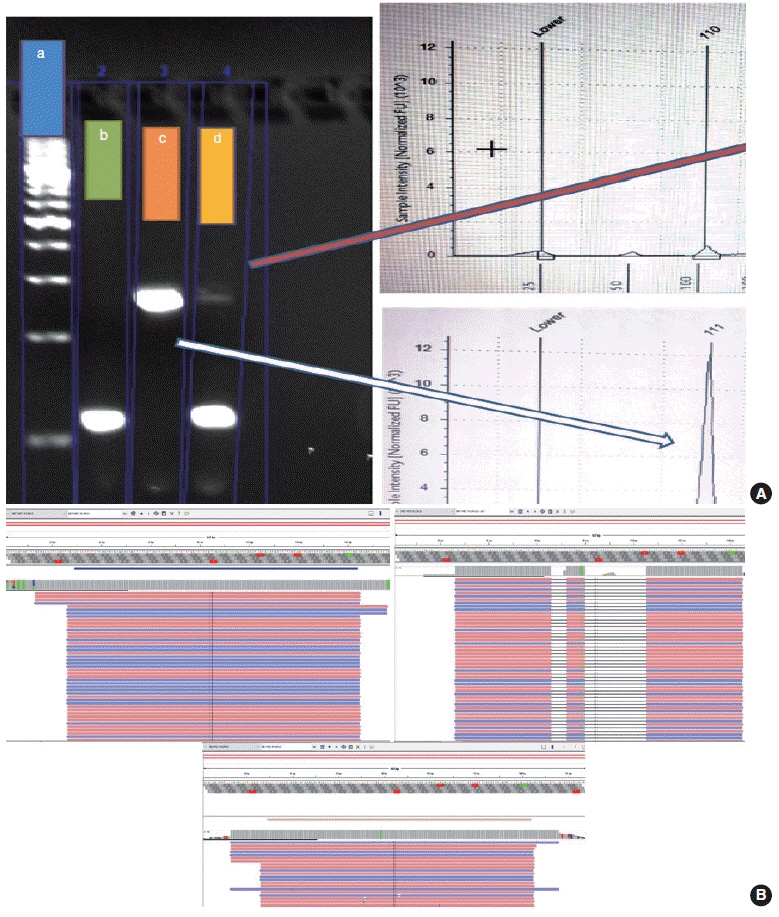
Thirteen cases of ΔMET14 mutation were detected on NGS using RNA-based sequencing. Considering NGS as a gold standard, the sizing assay using both gel and capillary electrophoresis that showed 100% specificity for both with concordance rates of 84.6% and 88.2% with NGS, respectively, were obtained.
Conclusions
Owing to the cost-effective nature and easy to use procedures, this assay will prove beneficial for small- and medium-sized laboratories where skilled technical personnel and NGS platforms are unavailable.
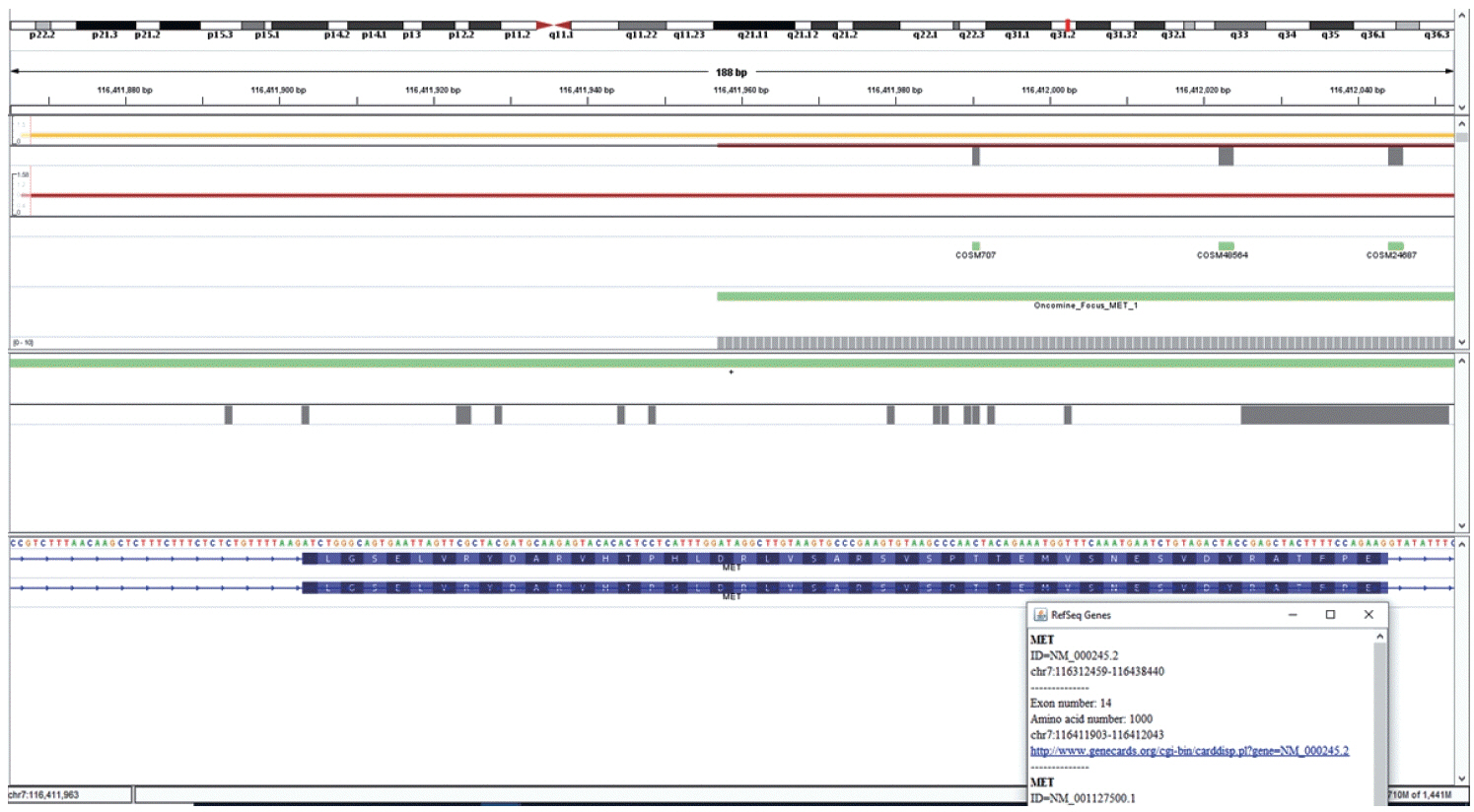
Figure
Reference
-
References
1. Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011; 3(1 Suppl):S7–19.
Article2. Saffroy R, Fallet V, Girard N, et al. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget. 2017; 8:42428–37.3. Tong JH, Yeung SF, Chan AW, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of nonsmall cell lung carcinoma with poor prognosis. Clin Cancer Res. 2016; 22:3048–56.4. Socinski MA, Pennell NA, Davies KD. MET exon 14 skipping mutations in non-small-cell lung cancer: an overview of biology, clinical outcomes, and testing considerations. JCO Precis Oncol. 2021; 5:PO.20.00516.5. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.17.00011.
Article6. Heist RS, Shim HS, Gingipally S, et al. MET exon 14 skipping in non-small cell lung cancer. Oncologist. 2016; 21:481–6.7. Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol. 2016; 11:1493–502.8. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–50.9. Wolf J, Han J, Nishio M, et al. GEOMETRY Mono-1: phase II, multicenter study of MET inhibitor capmatinib (INC280) in EGFR wt ,MET-dysregulated advanced NSCLC. J Thorac Oncol. 2017; 12(11 Suppl 1):S1578–9.10. Reungwetwattana T, Liang Y, Zhu V, Ou SI. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: the why, the how, the who, the unknown, and the inevitable. Lung Cancer. 2017; 103:27–37.
Article11. Buermans HP, den Dunnen JT. Next generation sequencing technology: advances and applications. Biochim Biophys Acta. 2014; 1842:1932–41.
Article12. Mehta A, Nathany S, Tripathi R, Sharma SK, Saifi M, Batra U. Non-amplification genetic alterations of HER2 gene in non-small cell lung carcinoma. J Clin Pathol. 2021; 74:106–10.13. Pilotto S, Gkountakos A, Carbognin L, Scarpa A, Tortora G, Bria E. MET exon 14 juxtamembrane splicing mutations: clinical and therapeutical perspectives for cancer therapy. Ann Transl Med. 2017; 5:2.14. Kim EK, Kim KA, Lee CY, et al. Molecular diagnostic assays and clinicopathologic implications of MET exon 14 skipping mutation in non-small-cell lung cancer. Clin Lung Cancer. 2019; 20:e123–32.15. O’Brien O, Wright MC, O’Brien C, et al. Cost-efficient and easy to perform PCR-based assay to identify Met exon 14 skipping in formalin-fixed paraffin-embedded (FFPE) non-small cell lung cancer (NSCLC) samples. Diagnostics (Basel). 2019; 9:13.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Clinical Impact of Capmatinib in the Treatment of Advanced Non–Small Cell Lung Cancer with MET Exon 14 Skipping Mutation or Gene Amplification
- Mutational Analysis of JAK1 Exons 10 and 13 in Non-small Cell Lung Cancers
- Does the efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor differ according to the type of EGFR mutation in non-small cell lung cancer?
- Analysis of TP53 Gene Mutations in the Korean Patients with Lung Cancer
- Comprison of p53 Mutation in Non Small Cell Lung Cancer between Young patients and Old Patients