Ann Surg Treat Res.
2024 Apr;106(4):195-202. 10.4174/astr.2024.106.4.195.
Comparative profiling by data-independent acquisition mass spectrometry reveals featured plasma proteins in breast cancer: a pilot study
- Affiliations
-
- 1Department of Surgery, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- 2Bertis R&D Division, Bertis Inc., Seongnam, Korea
- KMID: 2554478
- DOI: http://doi.org/10.4174/astr.2024.106.4.195
Abstract
- Purpose
Breast cancer is known to be influenced by genetic and environmental factors, and several susceptibility genes have been discovered. Still, the majority of genetic contributors remain unknown. We aimed to analyze the plasma proteome of breast cancer patients in comparison to healthy individuals to identify differences in protein expression profiles and discover novel biomarkers.
Methods
This pilot study was conducted using bioresources from Seoul National University Bundang Hospital’s Human Bioresource Center. Serum samples from 10 breast cancer patients and 10 healthy controls were obtained. Liquid chromatography-mass spectrometry analysis was performed to identify differentially expressed proteins.
Results
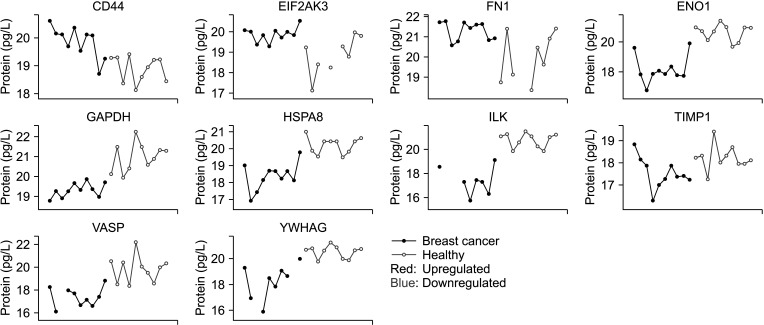
We identified 891 proteins; 805 were expressed in the breast cancer group and 882 in the control group. Gene set enrichment and differential expression analysis identified 30 upregulated and 100 downregulated proteins in breast cancer. Among these, 10 proteins were selected as potential biomarkers. Three proteins were upregulated in breast cancer patients, including cluster of differentiation 44, eukaryotic translation initiation factor 2-α kinase 3, and fibronectin 1. Seven proteins downregulated in breast cancer patients were also selected: glyceraldehyde-3-phosphate dehydrogenase, α-enolase, heat shock protein member 8, integrin‑linked kinase, tissue inhibitor of metalloproteinases-1, vasodilatorstimulated phosphoprotein, and 14-3-3 protein gamma. All proteins had been previously reported to be related to tumor development and progression.
Conclusion
The findings suggest that plasma proteome profiling can reveal potential diagnostic biomarkers for breast cancer and may contribute to early detection and personalized treatment strategies. A further validation study with a larger sample cohort of breast cancer patients is planned.
Keyword
Figure
Reference
-
1. Veronesi U, Boyle P, Goldhirsch A, Orecchia R, Viale G. Breast cancer. Lancet. 2005; 365:1727–1741. PMID: 15894099.2. Odle TG. Precision medicine in breast cancer. Radiol Technol. 2017; 88:401M–421M. PMID: 28298497.3. Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017; 551:92–94. PMID: 29059683.4. Schwenk JM, Igel U, Kato BS, Nicholson G, Karpe F, Uhlén M, et al. Comparative protein profiling of serum and plasma using an antibody suspension bead array approach. Proteomics. 2010; 10:532–540. PMID: 19953555.5. Huang Z, Ma L, Huang C, Li Q, Nice EC. Proteomic profiling of human plasma for cancer biomarker discovery. Proteomics. 2017; 17.6. Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017; 13:942. PMID: 28951502.7. Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020; 17:41–44. PMID: 31768060.8. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014; 13:2513–2526. PMID: 24942700.9. Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011; 39:D712–D717. PMID: 21071422.10. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. PMID: 10592173.11. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020; 48:D498–D503. PMID: 31691815.12. Martens M, Ammar A, Riutta A, Waagmeester A, Slenter DN, Hanspers K, et al. WikiPathways: connecting communities. Nucleic Acids Res. 2021; 49:D613–D621. PMID: 33211851.13. Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, et al. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci Signal. 2013; 6:rs4. PMID: 23443683.14. Kang UB, Ahn Y, Lee JW, Kim YH, Kim J, Yu MH, et al. Differential profiling of breast cancer plasma proteome by isotope-coded affinity tagging method reveals biotinidase as a breast cancer biomarker. BMC Cancer. 2010; 10:114. PMID: 20346108.15. Lumachi F, Basso SM. Serum tumor markers in patients with breast cancer. Expert Rev Anticancer Ther. 2004; 4:921–931. PMID: 15485325.16. Bayo J, Castaño MA, Rivera F, Navarro F. Analysis of blood markers for early breast cancer diagnosis. Clin Transl Oncol. 2018; 20:467–475. PMID: 28808872.17. Yao F, Yan C, Zhang Y, Shen L, Zhou D, Ni J. Identification of blood protein biomarkers for breast cancer staging by integrative transcriptome and proteome analyses. J Proteomics. 2021; 230:103991. PMID: 32971305.18. Olsson E, Honeth G, Bendahl PO, Saal LH, Gruvberger-Saal S, Ringnér M, et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011; 11:418. PMID: 21957977.19. Zhao C, Yin S, Dong Y, Guo X, Fan L, Ye M, et al. Autophagy-dependent EIF2AK3 activation compromises ursolic acid-induced apoptosis through upregulation of MCL1 in MCF-7 human breast cancer cells. Autophagy. 2013; 9:196–207. PMID: 23182854.20. Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016; 431:123–132. PMID: 27173027.21. Zhang XX, Luo JH, Wu LQ. FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front Genet. 2022; 13:913659. PMID: 36035176.22. Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ, et al. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol Med. 2015; 12:10–22. PMID: 25859407.23. Cancemi P, Buttacavoli M, Roz E, Feo S. Expression of alpha-enolase (ENO1), Myc promoter-binding protein-1 (MBP-1) and matrix metalloproteinases (MMP-2 and MMP-9) reflect the nature and aggressiveness of breast tumors. Int J Mol Sci. 2019; 20:3952. PMID: 31416219.24. Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006; 31:164–172. PMID: 16483782.25. Ying B, Xu W, Nie Y, Li Y. HSPA8 is a new biomarker of triple negative breast cancer related to prognosis and immune infiltration. Dis Markers. 2022; 2022:8446857. PMID: 36452344.26. Tsirtsaki K, Gkretsi V. The focal adhesion protein Integrin-Linked Kinase (ILK) as an important player in breast cancer pathogenesis. Cell Adh Migr. 2020; 14:204–213. PMID: 33043811.27. Würtz SO, Schrohl AS, Mouridsen H, Brünner N. TIMP-1 as a tumor marker in breast cancer: an update. Acta Oncol. 2008; 47:580–590. PMID: 18465326.28. Li K, Zhang J, Tian Y, He Y, Xu X, Pan W, et al. The Wnt/β-catenin/VASP positive feedback loop drives cell proliferation and migration in breast cancer. Oncogene. 2020; 39:2258–2274. PMID: 31831834.29. Hiraoka E, Mimae T, Ito M, Kadoya T, Miyata Y, Ito A, et al. Breast cancer cell motility is promoted by 14-3-3γ. Breast Cancer. 2019; 26:581–593. PMID: 30830684.30. Wang J, Pan X, Li J, Zhao J. TXNDC9 knockdown inhibits lung adenocarcinoma progression by targeting YWHAG. Mol Med Rep. 2022; 25:203. PMID: 35485284.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers
- Urinary Metabolites as Biomarkers for Diagnosis of Breast Cancer: A Preliminary Study
- A Validation Study of a Multiple Reaction Monitoring-Based Proteomic Assay to Diagnose Breast Cancer
- Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: A brief review
- Sporozoite proteome analysis of Cryptosporidium parvum by one-dimensional SDS-PAGE and liquid chromatography tandem mass spectrometry