Ann Lab Med.
2024 Mar;44(2):188-191. 10.3343/alm.2023.0163.
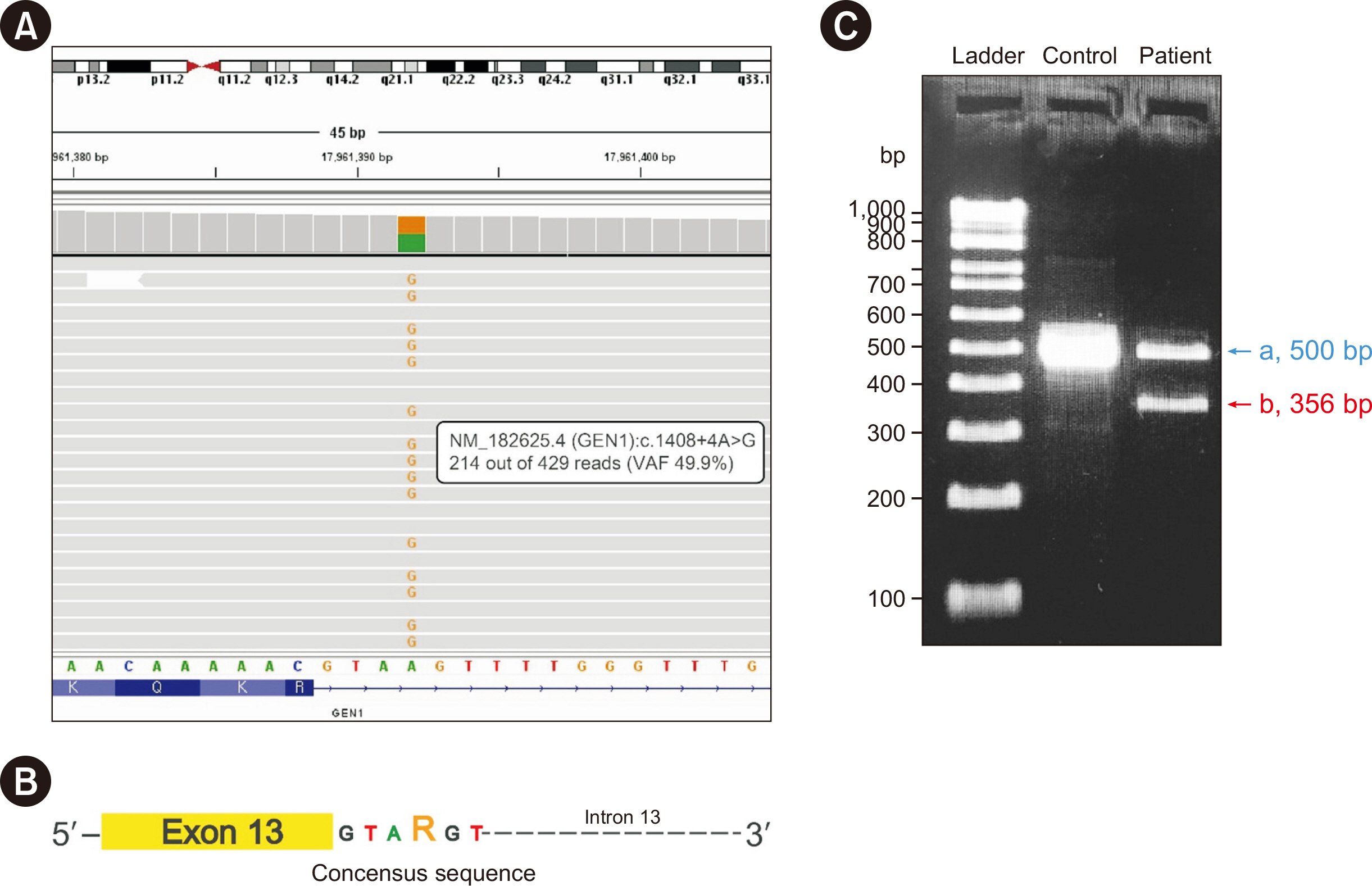
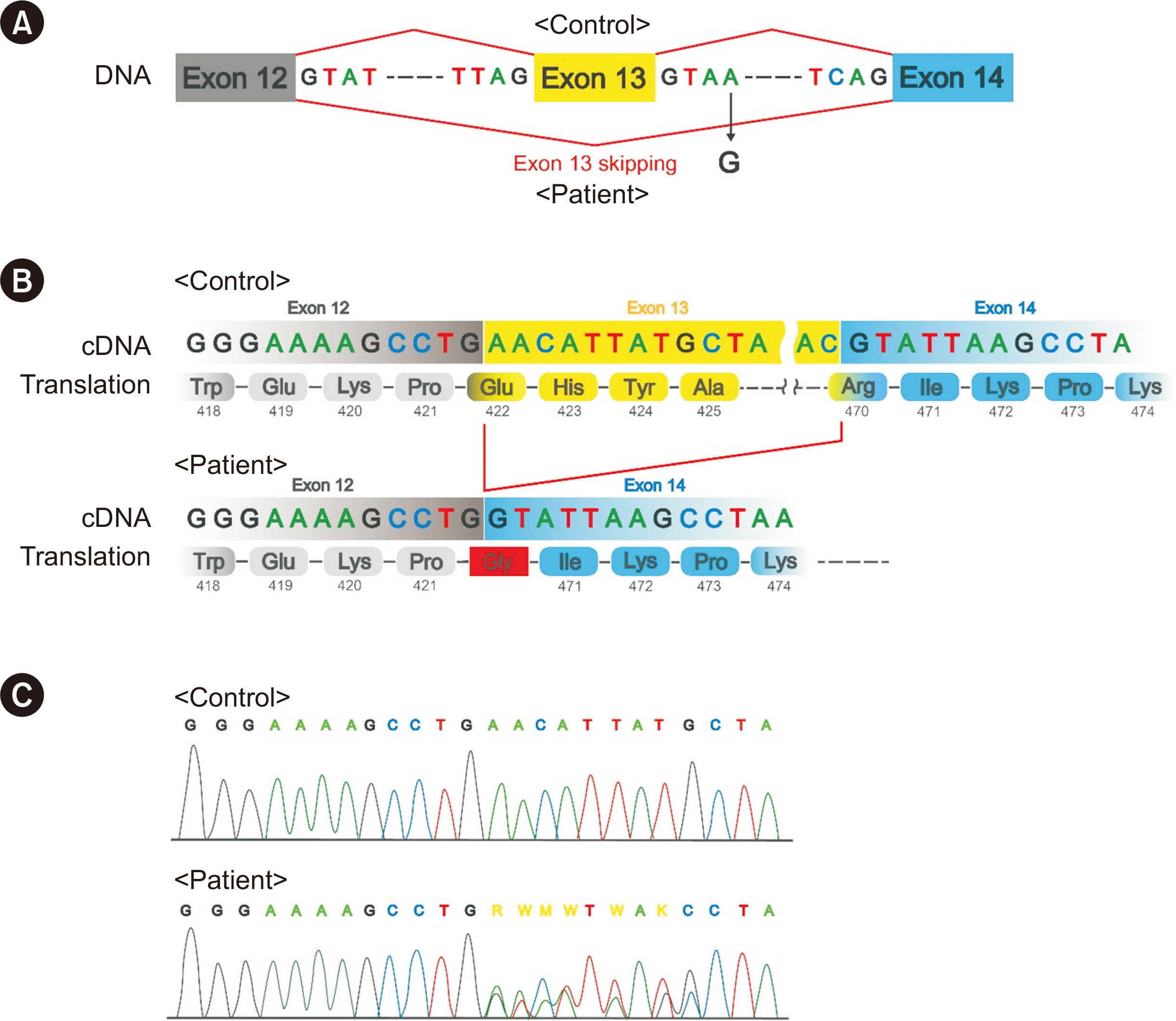
Reverse Transcription-PCR-based Sanger Sequencingconfirmed Exon-skipping Effect of a Novel GEN1 Intronic Variant (c.1408+4A>G)
- Affiliations
-
- 1Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Surgery, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2553392
- DOI: http://doi.org/10.3343/alm.2023.0163
Figure
Reference
-
1. National Cancer Control Institute. National Cancer Registration Program, Annual report of cancer statistics in Korea in 2020. http://www.ncc.re.kr. Updated on Dec 2022.2. Nemtsova MV, Kalinkin AI, Kuznetsova EB, Bure IV, Alekseeva EA, Bykov II, et al. 2020; Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci Rep. 10:504. DOI: 10.1038/s41598-020-57544-3. PMID: 31949278. PMCID: PMC6965114.
Article3. van der Post RS, Vogelaar IP, Carneiro F, Guilford P, Huntsman D, Hoogerbrugge N, et al. 2015; Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 52:361–74. DOI: 10.1136/jmedgenet-2015-103094. PMID: 25979631. PMCID: PMC4453626.
Article4. Lee SH, Princz LN, Klügel MF, Habermann B, Pfander B, Biertümpfel C. 2015; Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage. Elife. 4:e12256. DOI: 10.7554/eLife.12256. PMID: 26682650. PMCID: PMC5039027.
Article5. Li X, Heyer WD. 2008; Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18:99–113. DOI: 10.1038/cr.2008.1. PMID: 18166982. PMCID: PMC3087377.
Article6. Turnbull C, Hines S, Renwick A, Hughes D, Pernet D, Elliott A, et al. 2010; Mutation and association analysis of GEN1 in breast cancer susceptibility. Breast Cancer Res Treat. 124:283–8. DOI: 10.1007/s10549-010-0949-1. PMID: 20512659. PMCID: PMC3632835.
Article7. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. 2016; Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 375:443–53. DOI: 10.1056/NEJMoa1603144. PMID: 27433846. PMCID: PMC4986616.8. WHO Classification of Tumours Editorial Board. WHO Classification of Tumors: Digestive System Tumours. 5th ed. International Agency for Research on Cancer;Lyon, France: 2019.9. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article10. Gu H, Hong J, Lee W, Kim SB, Chun S, Min WK. 2022; RNA Sequencing for Elucidating an Intronic Variant of Uncertain Significance (SDHD c.314+3A>T) in Splicing Site Consensus Sequences. Ann Lab Med. 42:376–9. DOI: 10.3343/alm.2022.42.3.376. PMID: 34907111. PMCID: PMC8677484.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of ATM Mutations in Korean Siblings with Ataxia-Telangiectasia
- New Splicing Variants of the Murine Damaged DNA Binding 2
- Detection of RHD 1227G>A and 1222T>C Using PCR-Restriction Fragment Length Polymorphism
- A Case of Chronic Myeloid Leukemia with Micro BCR::ABL1 Rearrangement: Precaution in Reverse Transcription PCR to Prevent False Negativity
- Gene Therapy of Inherited Muscle Diseases