Cancer Res Treat.
2024 Jan;56(1):104-114. 10.4143/crt.2023.728.
Clinical Impact of Genomic and Pathway Alterations in Stage I EGFR-Mutant Lung Adenocarcinoma
- Affiliations
-
- 1Department of Pathology, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Changwon, Korea
- 2Department of Pathology, National Health Insurance Service Ilsan Hospital, Goyang, Korea
- 3Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2550327
- DOI: http://doi.org/10.4143/crt.2023.728
Abstract
- Purpose
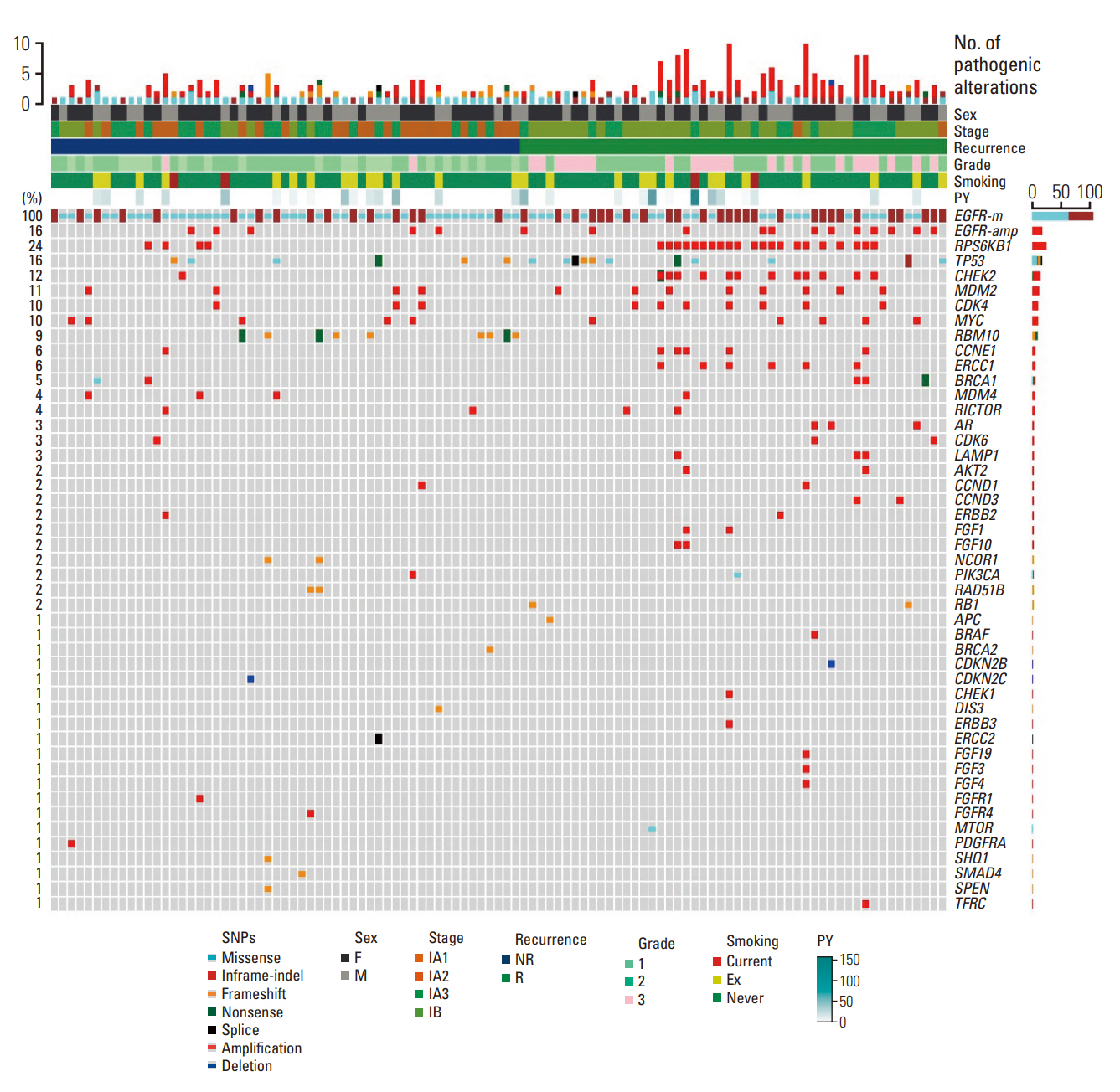
We investigated the clinical impact of genomic and pathway alterations in stage I epidermal growth factor receptor (EGFR)–mutant lung adenocarcinomas, which have a high recurrence rate despite complete surgical resection.
Materials and Methods
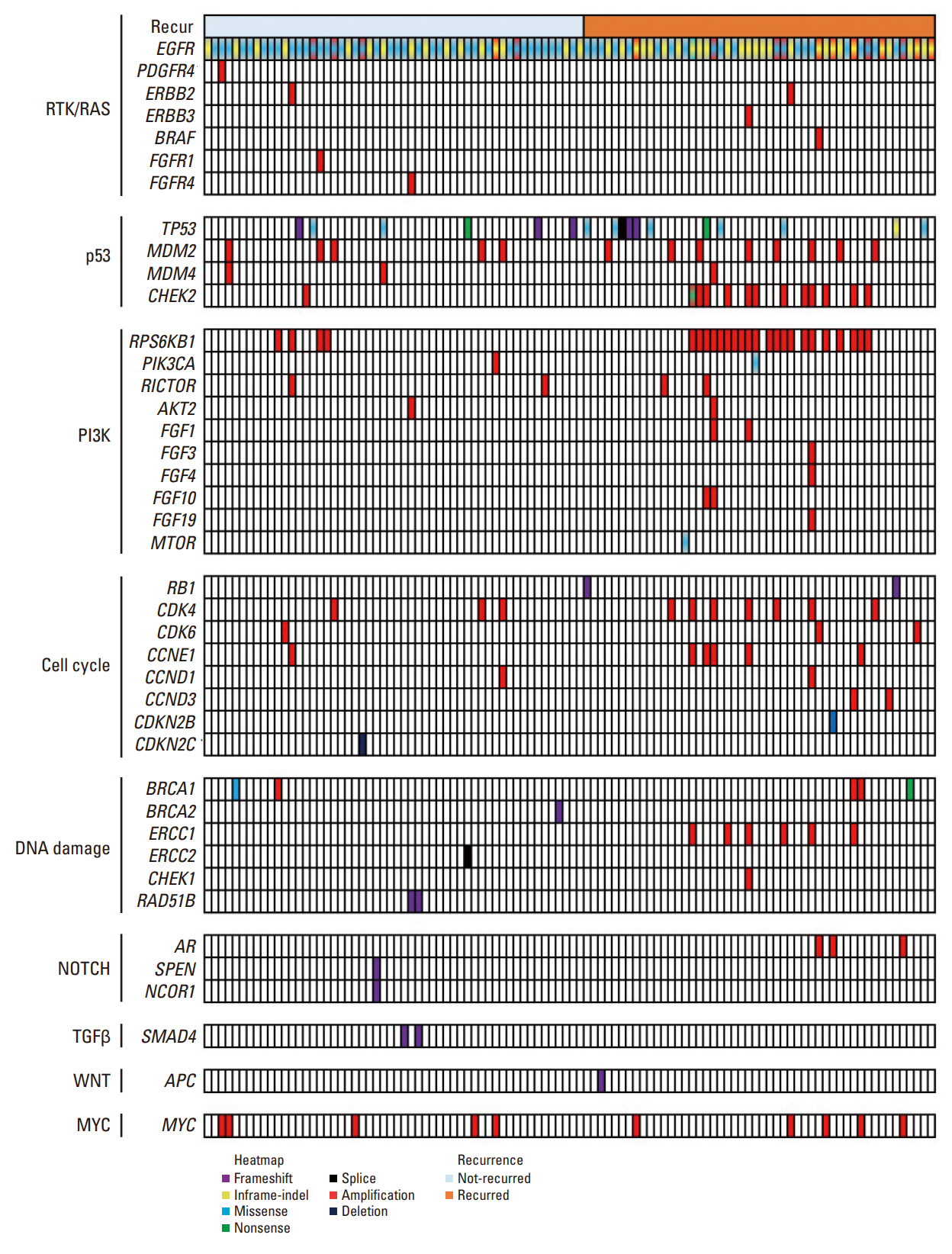
Out of the initial cohort of 257 patients with completely resected stage I EGFR-mutant lung adenocarcinoma, tumor samples from 105 patients were subjected to analysis using large-panel next-generation sequencing. We analyzed 11 canonical oncogenic pathways and determined the number of pathway alterations (NPA). Survival analyses were performed based on co-occurring alterations and NPA in three patient groups: all patients, patients with International Association for the Study of Lung Cancer (IASLC) pathology grade 2, and patients with recurrent tumors treated with EGFR–tyrosine kinase inhibitor (TKI).
Results
In the univariate analysis, pathological stage, IASLC grade, TP53 mutation, NPA, phosphoinositide 3-kinase pathway, p53 pathway, and cell cycle pathway exhibited significant associations with worse recurrence-free survival (RFS). Moreover, RPS6KB1 or EGFR amplifications were linked to a poorer RFS. Multivariate analysis revealed that pathologic stage, IASLC grade, and cell cycle pathway alteration were independent poor prognostic factors for RFS (p=0.002, p < 0.001, and p=0.006, respectively). In the grade 2 subgroup, higher NPA was independently associated with worse RFS (p=0.003). Additionally, in patients with recurrence treated with EGFR-TKIs, co-occurring TP53 mutations were linked to shorter progression-free survival (p=0.025).
Conclusion
Genomic and pathway alterations, particularly cell cycle alterations, high NPA, and TP53 mutations, were associated with worse clinical outcomes in stage I EGFR-mutant lung adenocarcinoma. These findings may have implications for risk stratification and the development of new therapeutic strategies in early-stage EGFR-mutant lung cancer patients.
Keyword
Figure
Reference
-
References
1. Chansky K, Detterbeck FC, Nicholson AG, Rusch VW, Vallieres E, Groome P, et al. The IASLC Lung Cancer Staging Project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2017; 12:1109–21.2. Jeon DS, Kim HC, Kim SH, Kim TJ, Kim HK, Moon MH, et al. Five-year overall survival and prognostic factors in patients with lung cancer: results from the Korean Association of Lung Cancer Registry (KALC-R) 2015. Cancer Res Treat. 2023; 55:103–11.3. Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2020; 383:1711–23.4. Liu SY, Bao H, Wang Q, Mao WM, Chen Y, Tong X, et al. Genomic signatures define three subtypes of EGFR-mutant stage II-III non-small-cell lung cancer with distinct adjuvant therapy outcomes. Nat Commun. 2021; 12:6450.5. Christopoulos P, Kirchner M, Roeper J, Saalfeld F, Janning M, Bozorgmehr F, et al. Risk stratification of EGFR(+) lung cancer diagnosed with panel-based next-generation sequencing. Lung Cancer. 2020; 148:105–12.6. Shukla S, Evans JR, Malik R, Feng FY, Dhanasekaran SM, Cao X, et al. Development of a RNA-Seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2017; 109:djw200.7. Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010; 70:36–45.8. Soltis AR, Bateman NW, Liu J, Nguyen T, Franks TJ, Zhang X, et al. Proteogenomic analysis of lung adenocarcinoma reveals tumor heterogeneity, survival determinants, and therapeutically relevant pathways. Cell Rep Med. 2022; 3:100819.9. Park E, Shim HS. Detection of targetable genetic alterations in Korean lung cancer patients: a comparison study of single-gene assays and targeted next-generation sequencing. Cancer Res Treat. 2020; 52:543–51.10. Moreira AL, Ocampo PS, Xia Y, Zhong H, Russell PA, Minami Y, et al. A grading system for invasive pulmonary sdenocarcinoma: a proposal from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2020; 15:1599–610.11. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.12. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018; 173:321–37.13. Zhou J, Sanchez-Vega F, Caso R, Tan KS, Brandt WS, Jones GD, et al. Analysis of tumor genomic pathway alterations using broad-panel next-generation sequencing in surgically resected lung adenocarcinoma. Clin Cancer Res. 2019; 25:7475–84.14. Kadota K, Kushida Y, Kagawa S, Ishikawa R, Ibuki E, Inoue K, et al. Cribriform subtype is an independent predictor of recurrence and survival after adjustment for the eighth edition of TNM staging system in patients with resected lung adenocarcinoma. J Thorac Oncol. 2019; 14:245–54.15. Ujiie H, Kadota K, Chaft JE, Buitrago D, Sima CS, Lee MC, et al. Solid predominant histologic subtype in resected stage I lung adenocarcinoma is an independent predictor of early, extrathoracic, multisite recurrence and of poor postrecurrence survival. J Clin Oncol. 2015; 33:2877–84.16. Nitadori J, Bograd AJ, Kadota K, Sima CS, Rizk NP, Morales EA, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst. 2013; 105:1212–20.17. Deng C, Zheng Q, Zhang Y, Jin Y, Shen X, Nie X, et al. Validation of the novel International Association for the Study of Lung Cancer grading system for invasive pulmonary adenocarcinoma and association with common driver mutations. J Thorac Oncol. 2021; 16:1684–93.18. Rokutan-Kurata M, Yoshizawa A, Ueno K, Nakajima N, Terada K, Hamaji M, et al. Validation study of the International Association for the Study of Lung Cancer histologic grading system of invasive lung adenocarcinoma. J Thorac Oncol. 2021; 16:1753–8.19. Park BJ, Woo W, Cha YJ, Shim HS, Yang YH, Moon DH, et al. Proposal of a revised International Association for the Study of Lung Cancer grading system in pulmonary non-mucinous adenocarcinoma: the importance of the lepidic proportion. Lung Cancer. 2023; 175:1–8.20. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019; 19:495–509.21. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. 2020; 15:1844–56.22. Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, et al. Impact of TP53 mutations on outcome in EGFR-mutated patients treated with first-line tyrosine kinase inhibitors. Clin Cancer Res. 2017; 23:2195–202.23. Kim IA, Hur JY, Kim HJ, Lee SA, Hwang JJ, Kim WS, et al. Targeted next-generation sequencing analysis predicts the recurrence in resected lung adenocarcinoma harboring EGFR mutations. Cancers (Basel). 2021; 13:3632.24. Jung HA, Lim J, Choi YL, Lee SH, Joung JG, Jeon YJ, et al. Clinical, pathologic, and molecular prognostic factors in patients with early-stage EGFR-mutant NSCLC. Clin Cancer Res. 2022; 28:4312–21.25. Nahar R, Zhai W, Zhang T, Takano A, Khng AJ, Lee YY, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018; 9:216.26. Vokes NI, Chambers E, Nguyen T, Coolidge A, Lydon CA, Le X, et al. Concurrent TP53 mutations facilitate resistance evolution in EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2022; 17:779–92.27. Perez-Tenorio G, Karlsson E, Waltersson MA, Olsson B, Holmlund B, Nordenskjold B, et al. Clinical potential of the mTOR targets S6K1 and S6K2 in breast cancer. Breast Cancer Res Treat. 2011; 128:713–23.28. Chen B, Yang L, Zhang R, Gan Y, Zhang W, Liu D, et al. Hyperphosphorylation of RPS6KB1, rather than overexpression, predicts worse prognosis in non-small cell lung cancer pati-ents. PLoS One. 2017; 12:e0182891.29. Kim M, Chung YS, Kim KA, Shim HS. Prognostic factors of acinar- or papillary-predominant adenocarcinoma of the lung. Lung Cancer. 2019; 137:129–35.30. Chaft JE, Rimner A, Weder W, Azzoli CG, Kris MG, Cascone T. Evolution of systemic therapy for stages I-III non-metastatic non-small-cell lung cancer. Nat Rev Clin Oncol. 2021; 18:547–57.31. Lengel HB, Connolly JG, Jones GD, Caso R, Zhou J, Sanchez-Vega F, et al. The emerging importance of tumor genomics in operable non-small cell lung cancer. Cancers (Basel). 2021; 13:3656.32. Ahn B, Yoon S, Kim D, Chun SM, Lee G, Kim HR, et al. Clinicopathologic and genomic features of high-grade pattern and their subclasses in lung adenocarcinoma. Lung Cancer. 2022; 170:176–84.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Small Cell Lung Cancer with Mutation of Epidermal Growth Factor Receptor in Patients with Lung Adenocarcinoma Resistant to Gefitinib
- Quercetin induces cell death by caspase-dependent and p38 MAPK pathway in EGFR mutant lung cancer cells
- A case of concomitant EGFR/ALK alteration against a mutated EGFR background in early-stage lung adenocarcinoma
- Expression of EGFR in Non-small Cell Lung Cancer and its Effects on Survival
- Clinical impact of pleural fluid carcinoembryonic antigen on therapeutic strategy and efficacy in lung adenocarcinoma patients with malignant pleural effusion