J Korean Diabetes.
2023 Dec;24(4):204-209. 10.4093/jkd.2023.24.4.204.
Precision Medicine in Treatment: Based on Multiomics or Clinical Data
- Affiliations
-
- 1School of Biological Sciences, Seoul National University, Seoul, Korea
- 2SNU Bioinformatics Institute, Seoul, Korea
- KMID: 2550160
- DOI: http://doi.org/10.4093/jkd.2023.24.4.204
Abstract
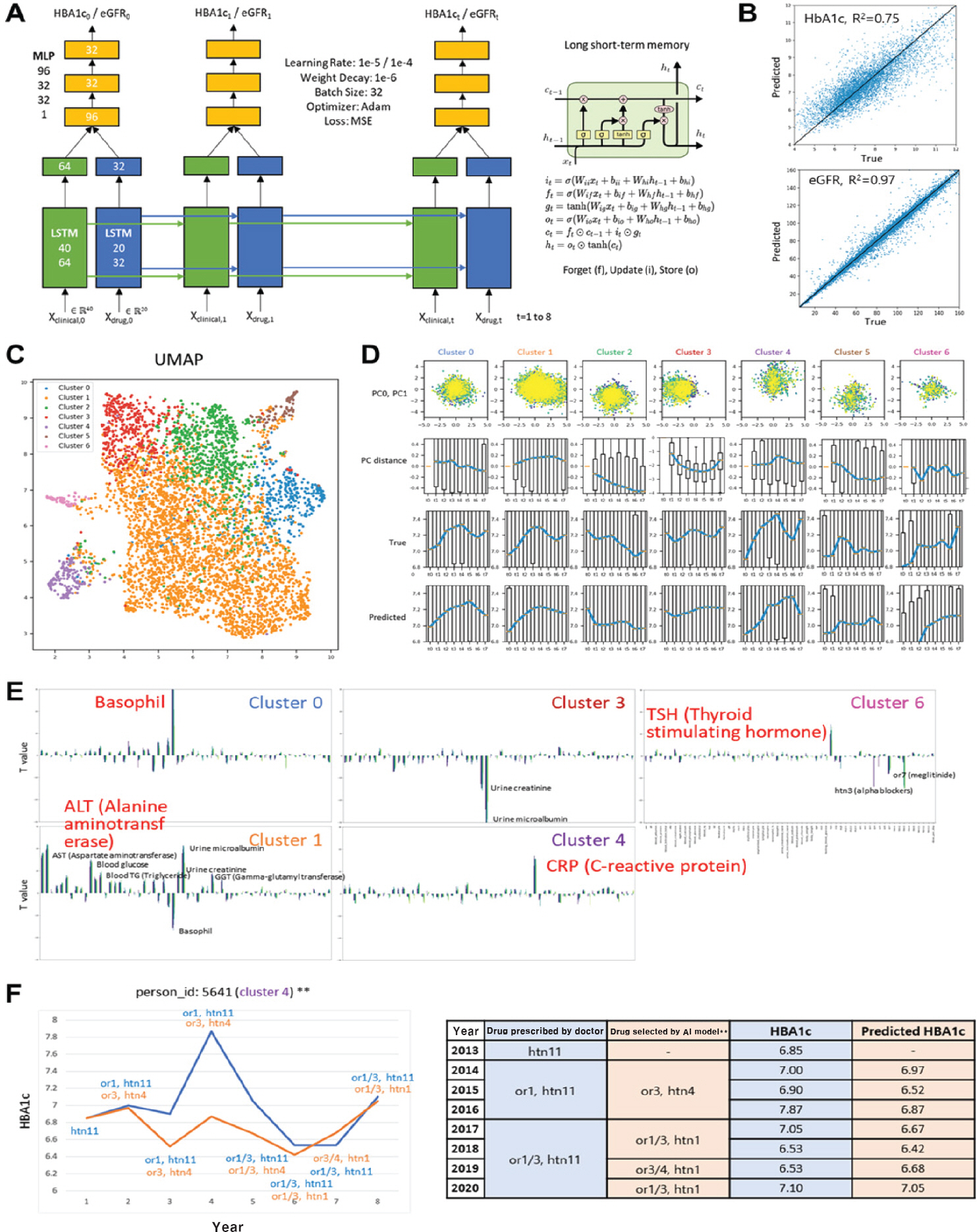
- Huge amounts of global data (genomic, epigenomic, transcriptomic, proteomic, and metabolomic) generated from a broad spectrum of specimens collected from human patients have accumulated in public repositories, together with electronic health records and drug treatment information. Accordingly, there is a need for bioinformatic methods that can effectively extract useful information for precision medicine. Here, we present two precision medicine approaches using multiomics data and clinical big data, respectively.
Figure
Reference
-
1.Mertins P., Mani DR., Ruggles KV., Gillette MA., Clauser KR., Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016. 534:55–62.2.Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014. 513:382–7.3.Zhang H., Liu T., Zhang Z., Payne SH., Zhang B., McDer-mott JE, et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016. 166:755–65.4.Mun DG., Bhin J., Kim S., Kim H., Jung JH., Jung Y, et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell. 2019. 35:111–24.e10.5.Hyeon DY., Nam D., Han Y., Kim DK., Kim G., Kim D, et al. Proteogenomic landscape of human pancreatic ductal adenocarcinoma in an Asian population reveals tumor cell-enriched and immune-rich subtypes. Nat Cancer. 2023. 4:290–307.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Multiomics Approach to Acromegaly: Unveiling Translational Insights for Precision Medicine
- Knowledge-guided artificial intelligence technologies for decoding complex multiomics interactions in cells
- Correspondence to editorial on “Multiomics profiling of buffy coat and plasma unveils etiology-specific signatures in hepatocellular carcinoma”
- Applying Precision Medicine in Clinical Practice
- Systems Biology of Human Microbiome for the Prediction of Personal Glycaemic Response