Diabetes Metab J.
2023 May;47(3):315-324. 10.4093/dmj.2022.0333.
Mitochondrial-Encoded Peptide MOTS-c, Diabetes, and Aging-Related Diseases
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea
- 2Leonard Davis School of Gerontology, University of Southern California, Los Angeles, CA, USA
- KMID: 2542508
- DOI: http://doi.org/10.4093/dmj.2022.0333
Abstract
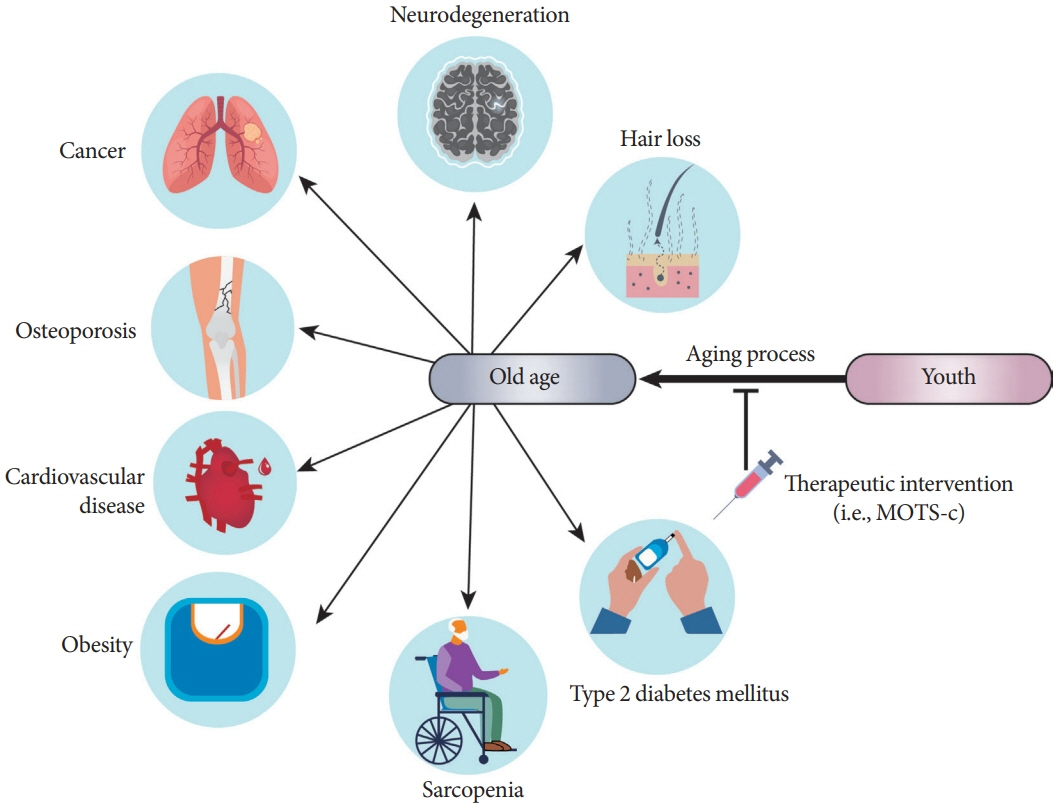
- Mitochondria are complex metabolic organelles with manifold pathophysiological implications in diabetes. Currently published mitochondrial-encoded peptides, which are expressed from the mitochondrial open reading frame of the 12S ribosomal RNA type-c (MOTS-c), 16S rRNA (humanin and short humanin like peptide 1-6 [SHLP1-6]), or small human mitochondrial open reading frame over serine tRNA (SHMOOSE) are associated with regulation of cellular metabolism and insulin action in age-related diseases, such as type 2 diabetes mellitus. This review focuses mainly on recent advances in MOTS-c research with regards to diabetes, including both type 1 and type 2. The emerging understanding of MOTS-c in diabetes may provide insight into the development of new therapies for diabetes and other age or senescence-related diseases.
Keyword
Figure
Reference
-
1. DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, et al. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015; 1:15019.
Article2. Katsarou A, Gudbjornsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers. 2017; 3:17016.
Article3. Faas MM, de Vos P. Mitochondrial function in immune cells in health and disease. Biochim Biophys Acta Mol Basis Dis. 2020; 1866:165845.4. Cho YM, Park KS, Lee HK. Genetic factors related to mitochondrial function and risk of diabetes mellitus. Diabetes Res Clin Pract. 2007; 77 Suppl 1:S172–7.5. Ukropcova B, Sereda O, de Jonge L, Bogacka I, Nguyen T, Xie H, et al. Family history of diabetes links impaired substrate switching and reduced mitochondrial content in skeletal muscle. Diabetes. 2007; 56:720–7.
Article6. Szendroedi J, Roden M. Mitochondrial fitness and insulin sensitivity in humans. Diabetologia. 2008; 51:2155–67.
Article7. Sivitz WI, Yorek MA. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal. 2010; 12:537–77.
Article8. Parish R, Petersen KF. Mitochondrial dysfunction and type 2 diabetes. Curr Diab Rep. 2005; 5:177–83.
Article9. Szendroedi J, Schmid AI, Meyerspeer M, Cervin C, Kacerovsky M, Smekal G, et al. Impaired mitochondrial function and insulin resistance of skeletal muscle in mitochondrial diabetes. Diabetes Care. 2009; 32:677–9.
Article10. Phielix E, Schrauwen-Hinderling VB, Mensink M, Lenaers E, Meex R, Hoeks J, et al. Lower intrinsic ADP-stimulated mitochondrial respiration underlies in vivo mitochondrial dysfunction in muscle of male type 2 diabetic patients. Diabetes. 2008; 57:2943–9.
Article11. Pagel-Langenickel I, Schwartz DR, Arena RA, Minerbi DC, Johnson DT, Waclawiw MA, et al. A discordance in rosiglitazone mediated insulin sensitization and skeletal muscle mitochondrial content/activity in type 2 diabetes mellitus. Am J Physiol Heart Circ Physiol. 2007; 293:H2659–66.
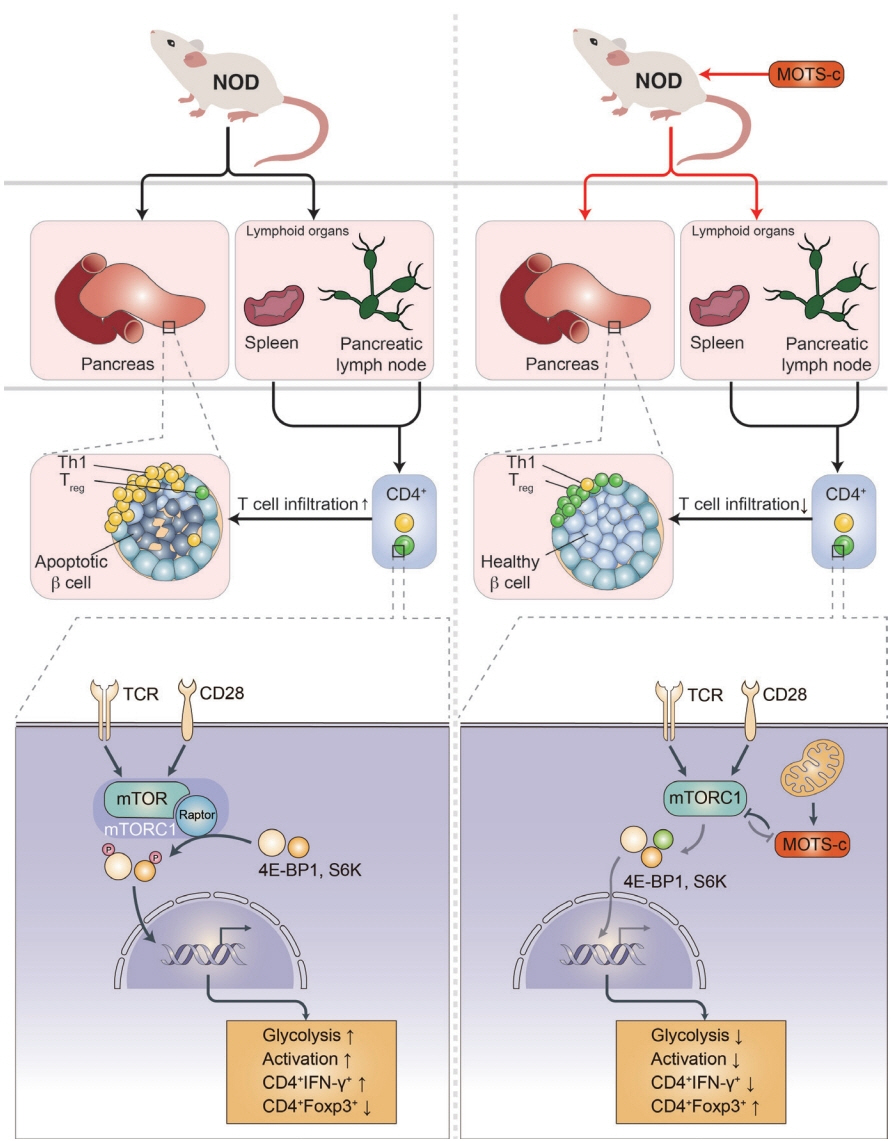
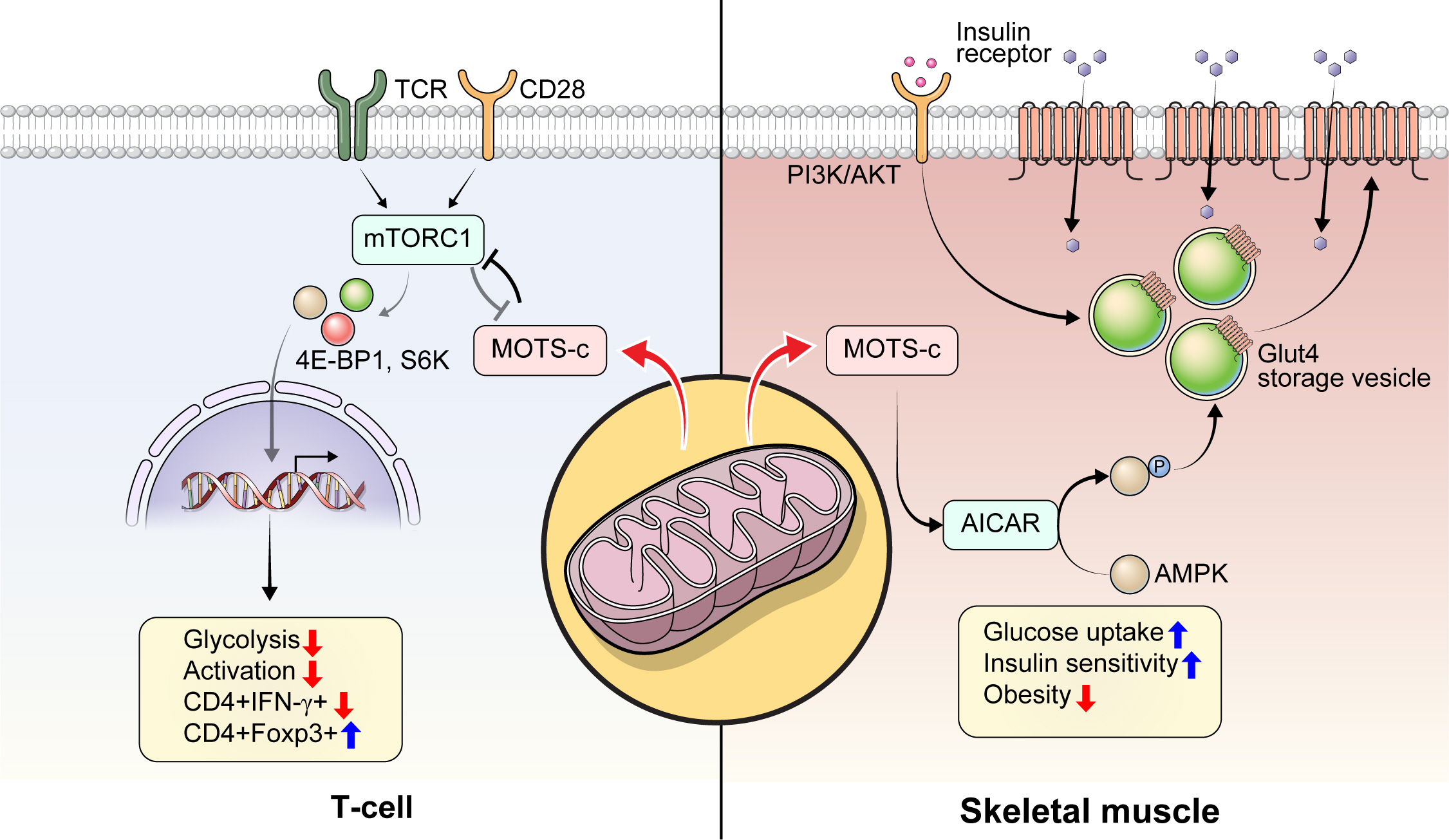
Article12. Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005; 115:3587–93.13. Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011; 14:528–36.14. Kong BS, Min SH, Lee C, Cho YM. Mitochondrial-encoded MOTS-c prevents pancreatic islet destruction in autoimmune diabetes. Cell Rep. 2021; 36:109447.
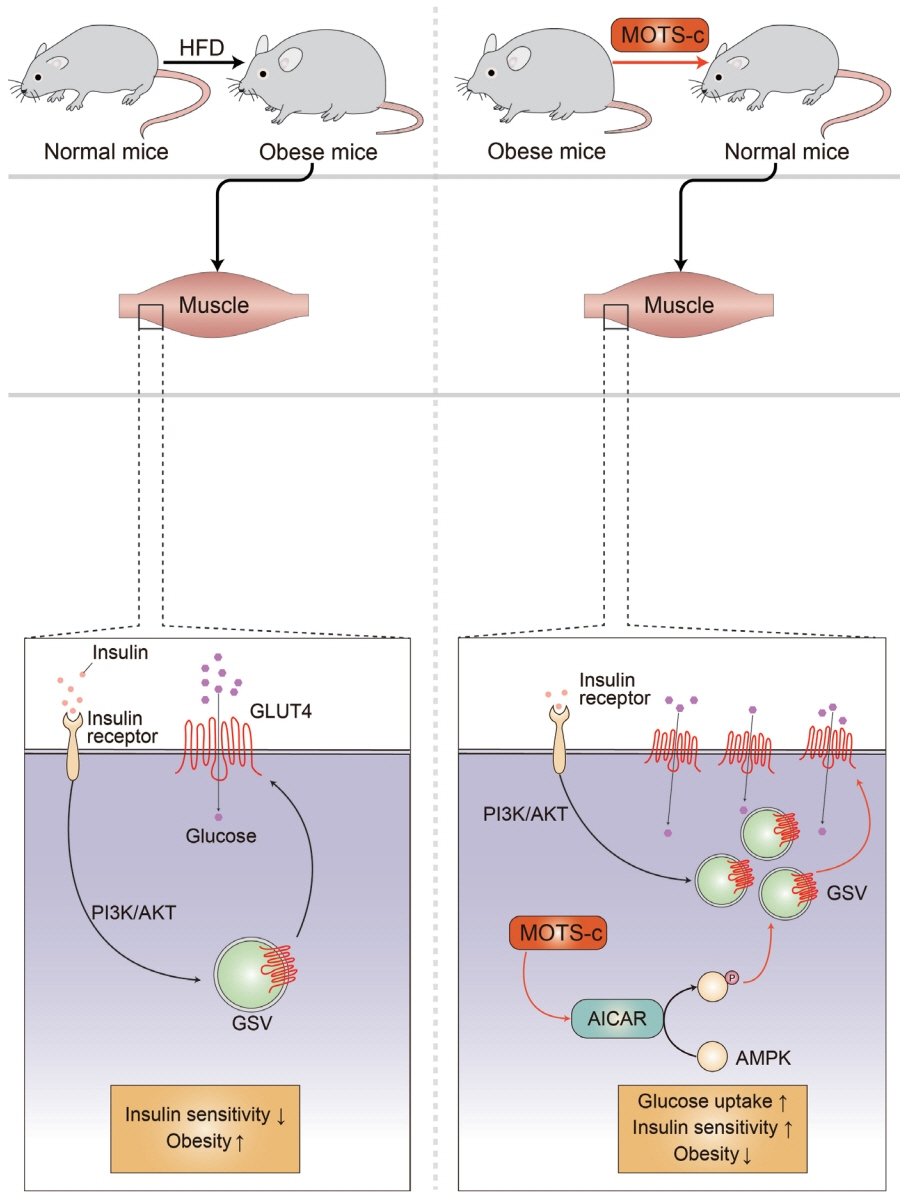
Article15. Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015; 21:443–54.
Article16. Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, et al. Naturally occurring mitochondrial-derived peptides are agedependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging (Albany NY). 2016; 8:796–809.
Article17. Miller B, Kim SJ, Kumagai H, Yen K, Cohen P. Mitochondriaderived peptides in aging and healthspan. J Clin Invest. 2022; 132:e158449.
Article18. Miller B, Kim SJ, Mehta HH, Cao K, Kumagai H, Thumaty N, et al. Mitochondrial DNA variation in Alzheimer’s disease reveals a unique microprotein called SHMOOSE. Mol Psychiatry. 2022; Sep. 21. [Epub]. htpps://doi.org/10.1038/s41380-022-01769-3.
Article19. Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-helix-mediated protein-protein interactions using designed molecules. Nat Chem. 2013; 5:161–73.
Article20. Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor design. J Am Chem Soc. 2011; 133:14220–3.
Article21. Edwards TA, Wilson AJ. Helix-mediated protein: protein interactions as targets for intervention using foldamers. Amino Acids. 2011; 41:743–54.22. Fairlie DP, West ML, Wong AK. Towards protein surface mimetics. Curr Med Chem. 1998; 5:29–62.
Article23. Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol. 2019; 15:635–50.
Article24. Monaco CM, Hughes MC, Ramos SV, Varah NE, Lamberz C, Rahman FA, et al. Correction to: altered mitochondrial bioenergetics and ultrastructure in the skeletal muscle of young adults with type 1 diabetes. Diabetologia. 2020; 63:887–8.
Article25. Padgett LE, Broniowska KA, Hansen PA, Corbett JA, Tse HM. The role of reactive oxygen species and proinflammatory cytokines in type 1 diabetes pathogenesis. Ann N Y Acad Sci. 2013; 1281:16–35.26. Corbett JA, Wang JL, Sweetland MA, Lancaster JR Jr, McDaniel ML. Interleukin 1 beta induces the formation of nitric oxide by beta-cells purified from rodent islets of Langerhans: evidence for the beta-cell as a source and site of action of nitric oxide. J Clin Invest. 1992; 90:2384–91.
Article27. Heitmeier MR, Scarim AL, Corbett JA. Prolonged STAT1 activation is associated with interferon-gamma priming for interleukin-1-induced inducible nitric-oxide synthase expression by islets of Langerhans. J Biol Chem. 1999; 274:29266–73.28. Chen J, Chernatynskaya AV, Li JW, Kimbrell MR, Cassidy RJ, Perry DJ, et al. T cells display mitochondria hyperpolarization in human type 1 diabetes. Sci Rep. 2017; 7:10835.
Article29. Hoang PT, Park P, Cobb LJ, Paharkova-Vatchkova V, Hakimi M, Cohen P, et al. The neurosurvival factor Humanin inhibits beta-cell apoptosis via signal transducer and activator of transcription 3 activation and delays and ameliorates diabetes in nonobese diabetic mice. Metabolism. 2010; 59:343–9.
Article30. Lytvyn Y, Wan J, Lai V, Cohen P, Cherney DZ. The effect of sex on humanin levels in healthy adults and patients with uncomplicated type 1 diabetes mellitus. Can J Physiol Pharmacol. 2015; 93:239–43.31. Rovira-Llopis S, Banuls C, Diaz-Morales N, Hernandez-Mijares A, Rocha M, Victor VM. Mitochondrial dynamics in type 2 diabetes: pathophysiological implications. Redox Biol. 2017; 11:637–45.
Article32. Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003; 300:1140–2.33. Lee HK, Song JH, Shin CS, Park DJ, Park KS, Lee KU, et al. Decreased mitochondrial DNA content in peripheral blood precedes the development of non-insulin-dependent diabetes mellitus. Diabetes Res Clin Pract. 1998; 42:161–7.34. Song J, Oh JY, Sung YA, Pak YK, Park KS, Lee HK. Peripheral blood mitochondrial DNA content is related to insulin sensitivity in offspring of type 2 diabetic patients. Diabetes Care. 2001; 24:865–9.35. Park KS, Song JH, Lee KU, Choi CS, Koh JJ, Shin CS, et al. Peripheral blood mitochondrial DNA content correlates with lipid oxidation rate during euglycemic clamps in healthy young men. Diabetes Res Clin Pract. 1999; 46:149–54.
Article36. Park KS, Lee KU, Song JH, Choi CS, Shin CS, Park DJ, et al. Peripheral blood mitochondrial DNA content is inversely correlated with insulin secretion during hyperglycemic clamp studies in healthy young men. Diabetes Res Clin Pract. 2001; 52:97–102.37. Constantin-Teodosiu D, Constantin D, Pelsers MM, Verdijk LB, van Loon L, Greenhaff PL. Mitochondrial DNA copy number associates with insulin sensitivity and aerobic capacity, and differs between sedentary, overweight middle-aged males with and without type 2 diabetes. Int J Obes (Lond). 2020; 44:929–36.
Article38. Zheng LD, Linarelli LE, Liu L, Wall SS, Greenawald MH, Seidel RW, et al. Insulin resistance is associated with epigenetic and genetic regulation of mitochondrial DNA in obese humans. Clin Epigenetics. 2015; 7:60.
Article39. Chung JY, Kim OY, Song J. Role of ketone bodies in diabetesinduced dementia: sirtuins, insulin resistance, synaptic plasticity, mitochondrial dysfunction, and neurotransmitter. Nutr Rev. 2022; 80:774–85.
Article40. Flamment M, Arvier M, Gallois Y, Simard G, Malthiery Y, Ritz P, et al. Fatty liver and insulin resistance in obese Zucker rats: no role for mitochondrial dysfunction. Biochimie. 2008; 90:1407–13.
Article41. Hu N, Dong M, Ren J. Hydrogen sulfide alleviates cardiac contractile dysfunction in an Akt2-knockout murine model of insulin resistance: role of mitochondrial injury and apoptosis. Am J Physiol Regul Integr Comp Physiol. 2014; 306:R761–71.42. Jeong EM, Chung J, Liu H, Go Y, Gladstein S, Farzaneh-Far A, et al. Role of mitochondrial oxidative stress in glucose tolerance, insulin resistance, and cardiac diastolic dysfunction. J Am Heart Assoc. 2016; 5:e003046.43. Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008; 102:401–14.44. Shikuma CM, Day LJ, Gerschenson M. Insulin resistance in the HIV-infected population: the potential role of mitochondrial dysfunction. Curr Drug Targets Infect Disord. 2005; 5:255–62.
Article45. Turner N, Robker RL. Developmental programming of obesity and insulin resistance: does mitochondrial dysfunction in oocytes play a role? Mol Hum Reprod. 2015; 21:23–30.46. Wang CH, Wei YH. Role of mitochondrial dysfunction and dysregulation of Ca2+ homeostasis in the pathophysiology of insulin resistance and type 2 diabetes. J Biomed Sci. 2017; 24:70.
Article47. Yokono K. Possible role of mitochondrial dysfunction in insulin resistance in the elderly. Nihon Rinsho. 2006; 64:39–44.48. Ahn CH, Choi EH, Kong BS, Cho YM. Effects of MOTS-c on the mitochondrial function of cells harboring 3243 A to G mutant mitochondrial DNA. Mol Biol Rep. 2020; 47:4029–35.
Article49. Baylan FA, Yarar E. Relationship between the mitochondriaderived peptide MOTS-c and insulin resistance in obstructive sleep apnea. Sleep Breath. 2021; 25:861–6.
Article50. Bhullar KS, Shang N, Kerek E, Wu K, Wu J. Mitofusion is required for MOTS-c induced GLUT4 translocation. Sci Rep. 2021; 11:14291.
Article51. Cataldo LR, Fernandez-Verdejo R, Santos JL, Galgani JE. Plasma MOTS-c levels are associated with insulin sensitivity in lean but not in obese individuals. J Investig Med. 2018; 66:1019–22.52. Du C, Zhang C, Wu W, Liang Y, Wang A, Wu S, et al. Circulating MOTS-c levels are decreased in obese male children and adolescents and associated with insulin resistance. Pediatr Diabetes. 2018; Apr. 25. [Epub]. htpps://doi.org/10.1111/pedi.12685.
Article53. Fuku N, Pareja-Galeano H, Zempo H, Alis R, Arai Y, Lucia A, et al. The mitochondrial-derived peptide MOTS-c: a player in exceptional longevity? Aging Cell. 2015; 14:921–3.
Article54. Kim KH, Son JM, Benayoun BA, Lee C. The mitochondrialencoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress. Cell Metab. 2018; 28:516–24.
Article55. Sequeira IR, Woodhead JST, Chan A, D’Souza RF, Wan J, Hollingsworth KG, et al. Plasma mitochondrial derived peptides MOTS-c and SHLP2 positively associate with android and liver fat in people without diabetes. Biochim Biophys Acta Gen Subj. 2021; 1865:129991.
Article56. Yin Y, Pan Y, He J, Zhong H, Wu Y, Ji C, et al. The mitochondrial-derived peptide MOTS-c relieves hyperglycemia and insulin resistance in gestational diabetes mellitus. Pharmacol Res. 2022; 175:105987.
Article57. Zempo H, Kim SJ, Fuku N, Nishida Y, Higaki Y, Wan J, et al. A pro-diabetogenic mtDNA polymorphism in the mitochondrial-derived peptide, MOTS-c. Aging (Albany NY). 2021; 13:1692–717.
Article58. Ramanjaneya M, Bettahi I, Jerobin J, Chandra P, Abi Khalil C, Skarulis M, et al. Mitochondrial-derived peptides are down regulated in diabetes subjects. Front Endocrinol (Lausanne). 2019; 10:331.
Article59. Crispim D, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m. 4216T > C and m.4917A > G variants. Ann Hum Genet. 2006; 70(Pt 4):488–95.60. Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007; 80:407–15.
Article61. Qin Q, Delrio S, Wan J, Jay Widmer R, Cohen P, Lerman LO, et al. Downregulation of circulating MOTS-c levels in patients with coronary endothelial dysfunction. Int J Cardiol. 2018; 254:23–7.
Article62. D’Souza RF, Woodhead JS, Hedges CP, Zeng N, Wan J, Kumagai H, et al. Increased expression of the mitochondrial derived peptide, MOTS-c, in skeletal muscle of healthy aging men is associated with myofiber composition. Aging (Albany NY). 2020; 12:5244–58.
Article63. Lu H, Wei M, Zhai Y, Li Q, Ye Z, Wang L, et al. MOTS-c peptide regulates adipose homeostasis to prevent ovariectomy-induced metabolic dysfunction. J Mol Med (Berl). 2019; 97:473–85.
Article64. Ming W, Lu G, Xin S, Huanyu L, Yinghao J, Xiaoying L, et al. Mitochondria related peptide MOTS-c suppresses ovariectomy-induced bone loss via AMPK activation. Biochem Biophys Res Commun. 2016; 476:412–9.
Article65. Curtis R, Geesaman BJ, DiStefano PS. Ageing and metabolism: drug discovery opportunities. Nat Rev Drug Discov. 2005; 4:569–80.66. Dao T, Green AE, Kim YA, Bae SJ, Ha KT, Gariani K, et al. Sarcopenia and muscle aging: a brief overview. Endocrinol Metab (Seoul). 2020; 35:716–32.
Article67. Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K, Pestka S. The majority of cDNA clones with strong positive signals for the interferon-induction-specific sequences resemble mitochondrial ribosomal RNA genes. Biochem Biophys Res Commun. 1983; 114:670–6.
Article68. Lei Y, Guerra Martinez C, Torres-Odio S, Bell SL, Birdwell CE, Bryant JD, et al. Elevated type I interferon responses potentiate metabolic dysfunction, inflammation, and accelerated aging in mtDNA mutator mice. Sci Adv. 2021; 7:eabe7548.69. Roy ER, Wang B, Wan YW, Chiu G, Cole A, Yin Z, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest. 2020; 130:1912–30.
Article70. Lasry A, Ben-Neriah Y. Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol. 2015; 36:217–28.
Article71. Kim SJ, Mehta HH, Wan J, Kuehnemann C, Chen J, Hu JF, et al. Mitochondrial peptides modulate mitochondrial function during cellular senescence. Aging (Albany NY). 2018; 10:1239–56.
Article72. Reynolds JC, Lai RW, Woodhead JS, Joly JH, Mitchell CJ, Cameron-Smith D, et al. MOTS-c is an exercise-induced mitochondrial-encoded regulator of age-dependent physical decline and muscle homeostasis. Nat Commun. 2021; 12:470.73. Yang B, Yu Q, Chang B, Guo Q, Xu S, Yi X, et al. MOTS-c interacts synergistically with exercise intervention to regulate PGC1α expression, attenuate insulin resistance and enhance glucose metabolism in mice via AMPK signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2021; 1867:166126.
Article74. Hyatt JK. MOTS-c increases in skeletal muscle following longterm physical activity and improves acute exercise performance after a single dose. Physiol Rep. 2022; 10:e15377.
Article75. Kang GM, Min SH, Lee CH, Kim JY, Lim HS, Choi MJ, et al. Mitohormesis in hypothalamic POMC neurons mediates regular exercise-induced high-turnover metabolism. Cell Metab. 2021; 33:334–49.
Article76. Yoon TK, Lee CH, Kwon O, Kim MS. Exercise, mitohormesis, and mitochondrial ORF of the 12S rRNA type-c (MOTS-c). Diabetes Metab J. 2022; 46:402–13.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Exercise, Mitohormesis, and Mitochondrial ORF of the 12S rRNA Type-C (MOTS-c)
- Beneficial Effects of Low-Grade Mitochondrial Stress on Metabolic Diseases and Aging
- Mitochondrial Gene Therapy
- Glucagon-Like Peptide-1 Increases Mitochondrial Biogenesis and Function in INS-1 Rat Insulinoma Cells
- Mechanism of Weight Loss and Diabetes Remission after Bariatric/Metabolic Surgery