J Pathol Transl Med.
2023 Jan;57(1):43-51. 10.4132/jptm.2022.12.12.
Single-cell and spatial sequencing application in pathology
- Affiliations
-
- 1Department of Microbiology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2Precision Medicine Research Center/Integrated Research Center for Genome Polymorphism, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Biomedicine & Health Sciences, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 4Department of Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 5Cancer Evolution Research Center, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2539035
- DOI: http://doi.org/10.4132/jptm.2022.12.12
Abstract
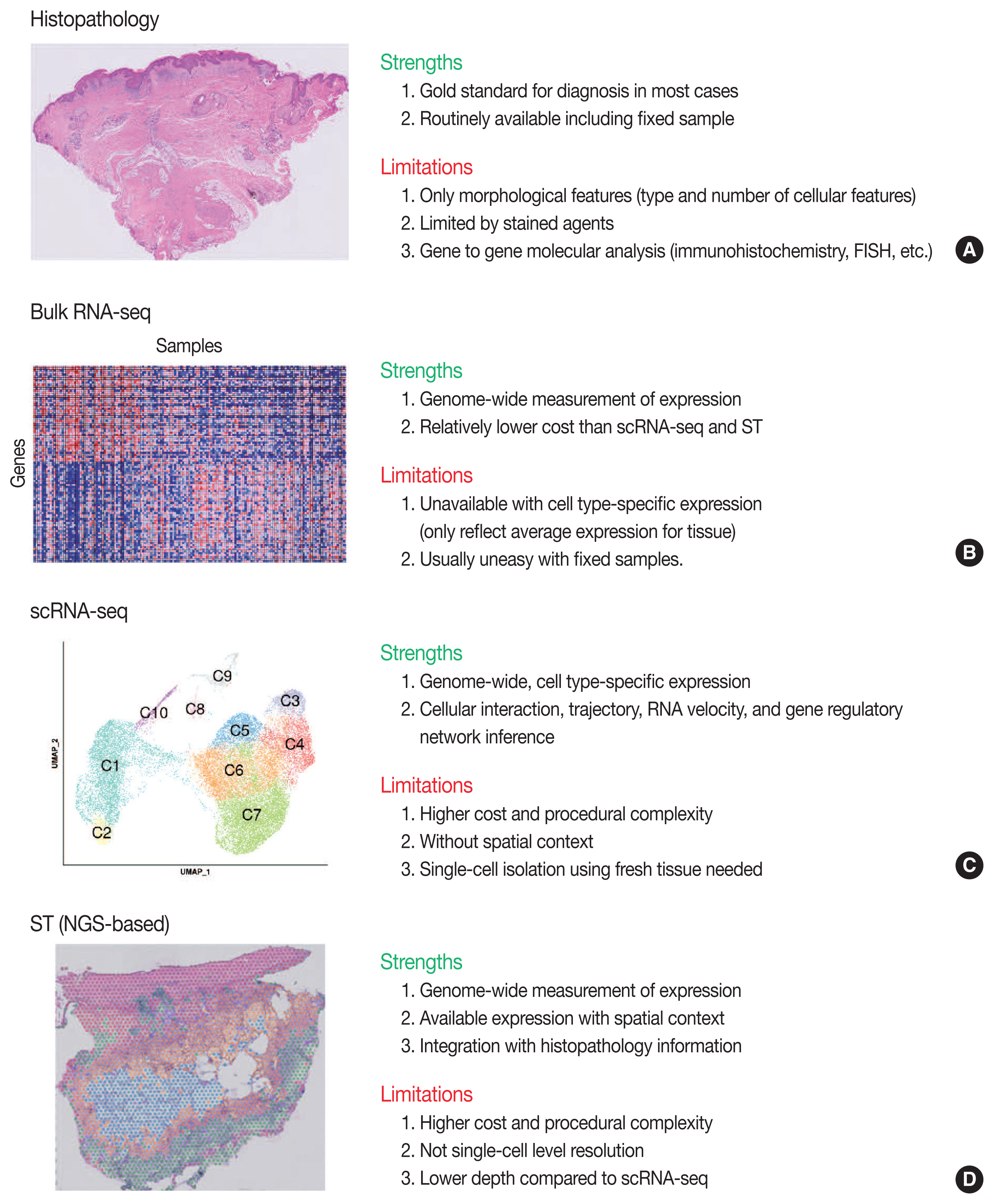
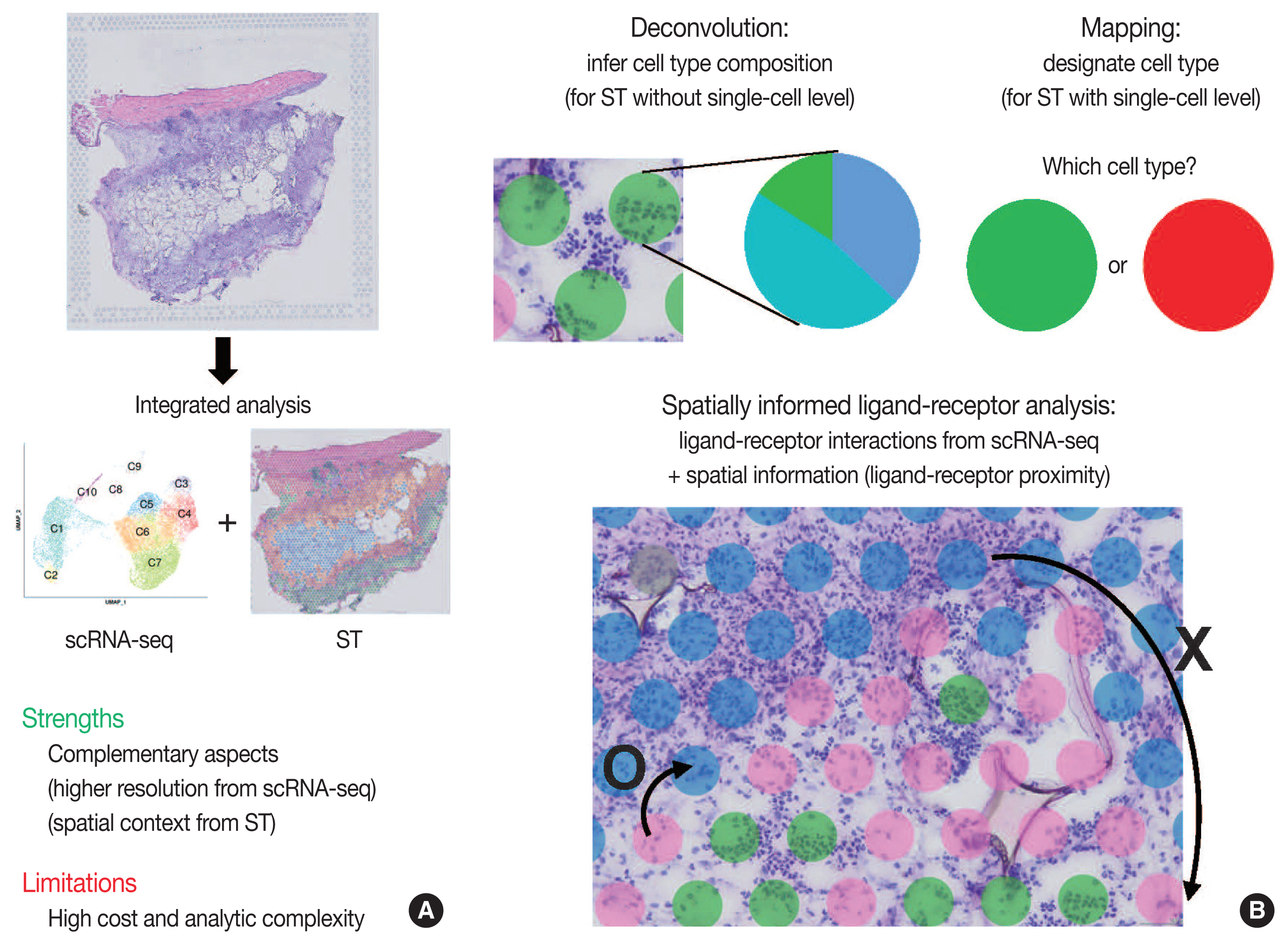
- Traditionally, diagnostic pathology uses histology representing structural alterations in a disease’s cells and tissues. In many cases, however, it is supplemented by other morphology-based methods such as immunohistochemistry and fluorescent in situ hybridization. Single-cell RNA sequencing (scRNA-seq) is one of the strategies that may help tackle the heterogeneous cells in a disease, but it does not usually provide histologic information. Spatial sequencing is designed to assign cell types, subtypes, or states according to the mRNA expression on a histological section by RNA sequencing. It can provide mRNA expressions not only of diseased cells, such as cancer cells but also of stromal cells, such as immune cells, fibroblasts, and vascular cells. In this review, we studied current methods of spatial transcriptome sequencing based on their technical backgrounds, tissue preparation, and analytic procedures. With the pathology examples, useful recommendations for pathologists who are just getting started to use spatial sequencing analysis in research are provided here. In addition, leveraging spatial sequencing by integration with scRNA-seq is reviewed. With the advantages of simultaneous histologic and single-cell information, spatial sequencing may give a molecular basis for pathological diagnosis, improve our understanding of diseases, and have potential clinical applications in prognostics and diagnostic pathology.
Figure
Reference
-
References
1. Stewart BJ, Clatworthy MR. Applying single-cell technologies to clinical pathology: progress in nephropathology. J Pathol. 2020; 250:693–704.
Article2. Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018; 50:1–14.
Article3. Liu B, Li Y, Zhang L. Analysis and visualization of spatial transcriptomic data. Front Genet. 2021; 12:785290.
Article4. Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021; 22:627–44.
Article5. Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009; 6:377–82.
Article6. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017; 8:14049.
Article7. Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019; 10:317.
Article8. Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019; 15:e8746.
Article9. Andrews TS, Kiselev VY, McCarthy D, Hemberg M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat Protoc. 2021; 16:1–9.
Article10. Kashima Y, Sakamoto Y, Kaneko K, Seki M, Suzuki Y, Suzuki A. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med. 2020; 52:1419–27.
Article11. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018; 18:35–45.
Article12. Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017; 65:631–43.
Article13. Rao A, Barkley D, Franca GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021; 596:211–20.
Article14. Lewis SM, Asselin-Labat ML, Nguyen Q, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021; 18:997–1012.
Article15. Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022; 14:68.
Article16. Moor AE, Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol. 2017; 46:126–33.
Article17. Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y. Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med. 2022; 12:e694.
Article18. Haque A, Engel J, Teichmann SA, Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017; 9:75.
Article19. Denisenko E, Guo BB, Jones M, et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020; 21:130.
Article20. Lafzi A, Moutinho C, Picelli S, Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc. 2018; 13:2742–57.
Article21. Slyper M, Porter CBM, Ashenberg O, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020; 26:792–802.
Article22. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021; 184:3573–87.
Article23. Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019; 20:163–72.
Article24. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014; 32:381–6.
Article25. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018; 560:494–8.
Article26. Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods. 2020; 17:147–54.
Article27. Skok Gibbs C, Jackson CA, Saldi GA, et al. High-performance single-cell gene regulatory network inference at scale: the Inferelator 3.0. Bioinformatics. 2022; 38:2519–28.
Article28. Zhuang X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat Methods. 2021; 18:18–22.
Article29. Larsson L, Frisen J, Lundeberg J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat Methods. 2021; 18:15–8.
Article30. Villacampa EG, Larsson L, Mirzazadeh R, et al. Genome-wide spatial expression profiling in formalin-fixed tissues. Cell Genomics. 2021; 1:100065.
Article31. Elosua-Bayes M, Nieto P, Mereu E, Gut I, Heyn H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021; 49:e50.
Article32. Bergenstrahle L, He B, Bergenstrahle J, et al. Super-resolved spatial transcriptomics by deep data fusion. Nat Biotechnol. 2022; 40:476–9.
Article33. Qian X, Harris KD, Hauling T, et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat Methods. 2020; 17:101–6.
Article34. Arora R, Cao C, Kumar M, et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Preprint at: https://doi.org/10.1101/2022.09.04.505581 . 2022.
Article35. Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021; 22:71–88.
Article36. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020; 15:1484–506.
Article37. Dries R, Zhu Q, Dong R, et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021; 22:78.
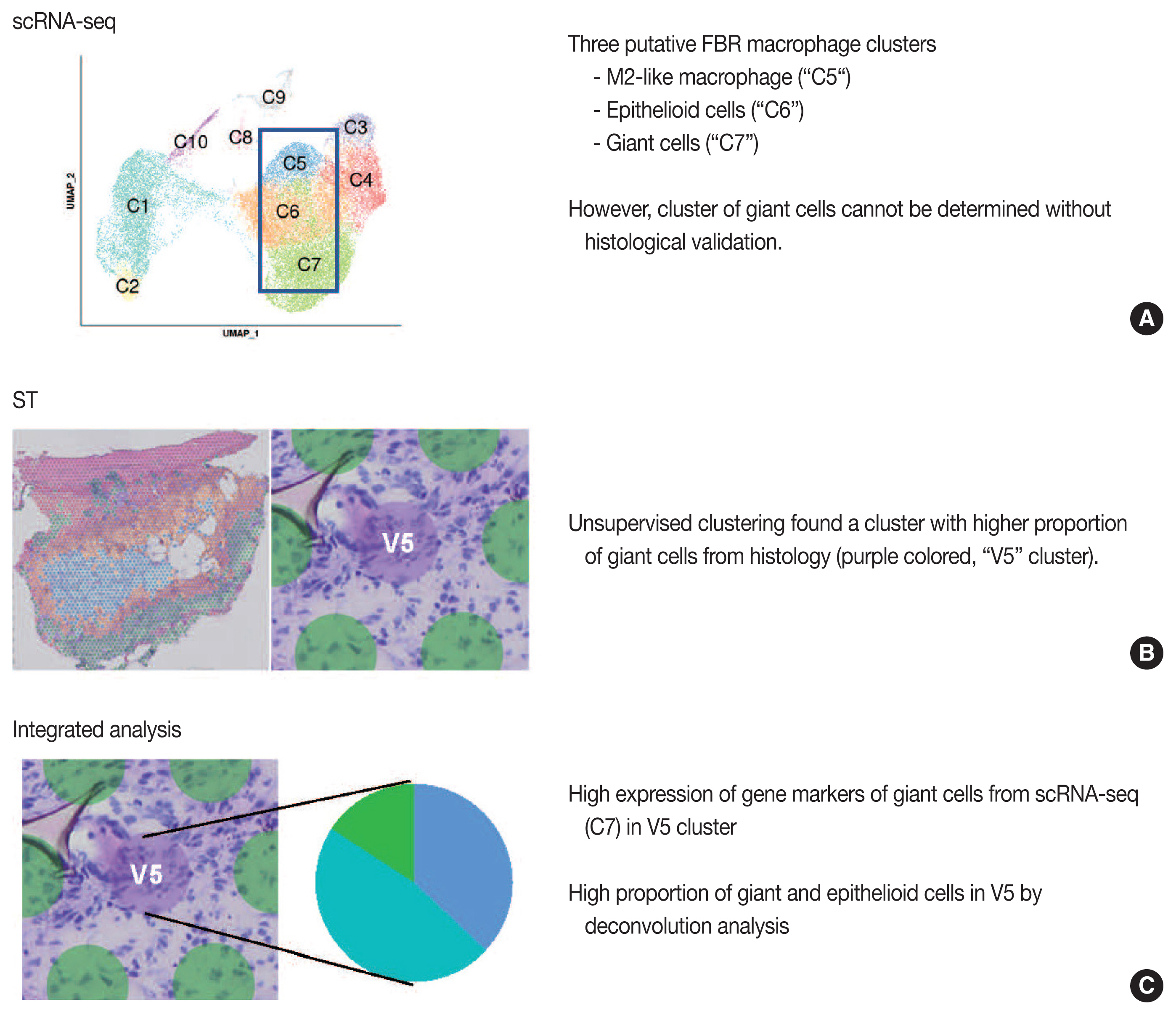
Article38. Kim YS, Shin S, Choi EJ, et al. Different molecular features of epithelioid and giant cells in foreign body reaction identified by single-cell RNA sequencing. J Invest Dermatol. 2022; 142:3232–42.
Article39. Ji AL, Rubin AJ, Thrane K, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020; 182:497–514.
Article40. Moncada R, Barkley D, Wagner F, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020; 38:333–42.
Article41. Argelaguet R, Clark SJ, Mohammed H, et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019; 576:487–91.
Article42. Abascal F, Harvey LM, Mitchell E, et al. Somatic mutation landscapes at single-molecule resolution. Nature. 2021; 593:405–10.
Article43. Lee J, Hyeon DY, Hwang D. Single-cell multiomics: technologies and data analysis methods. Exp Mol Med. 2020; 52:1428–42.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Applications of Single-Cell Omics Technologies for Induced Pluripotent Stem Cell-Based Cardiovascular Research
- Strategy of Patient-Specific Therapeutics in Cardiovascular Disease Through Single-Cell RNA Sequencing
- What Single Cell RNA Sequencing Has Taught Us about Chronic Obstructive Pulmonary Disease
- Application of Next Generation Sequencing in Laboratory Medicine
- Single-Cell Sequencing in Cancer: Recent Applications to Immunogenomics and Multi-omics Tools