J Korean Med Sci.
2022 Jul;37(26):e205. 10.3346/jkms.2022.37.e205.
Perceived Risk of Re-Identification in OMOP-CDM Database: A CrossSectional Survey
- Affiliations
-
- 1Department of Information Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Biomedical Systems Informatics, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Big Data Science, Halla University, Wonju, Korea
- 4UPS Data Corporation, Seoul, Korea
- KMID: 2531037
- DOI: http://doi.org/10.3346/jkms.2022.37.e205
Abstract
- Background
The advancement of information technology has immensely increased the quality and volume of health data. This has led to an increase in observational study, as well as to the threat of privacy invasion. Recently, a distributed research network based on the common data model (CDM) has emerged, enabling collaborative international medical research without sharing patient-level data. Although the CDM database for each institution is built inside a firewall, the risk of re-identification requires management. Hence, this study aims to elucidate the perceptions CDM users have towards CDM and risk management for re-identification.
Methods
The survey, targeted to answer specific in-depth questions on CDM, was conducted from October to November 2020. We targeted well-experienced researchers who actively use CDM. Basic statistics (total number and percent) were computed for all covariates.
Results
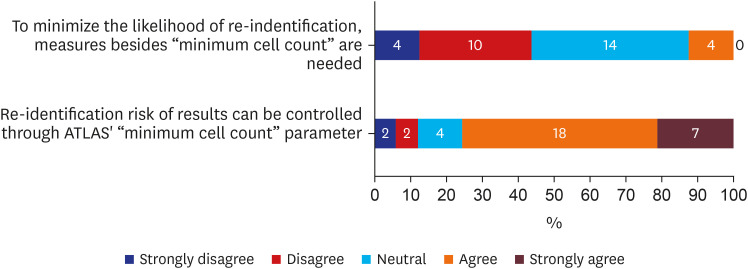
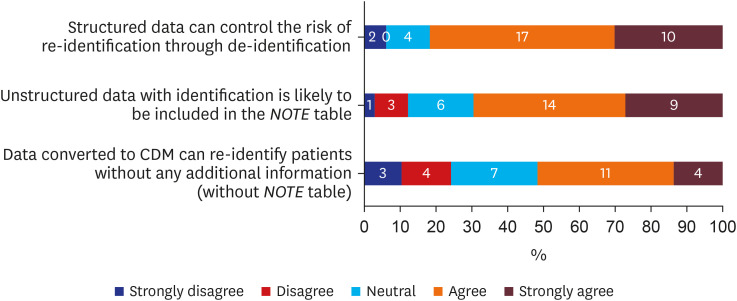
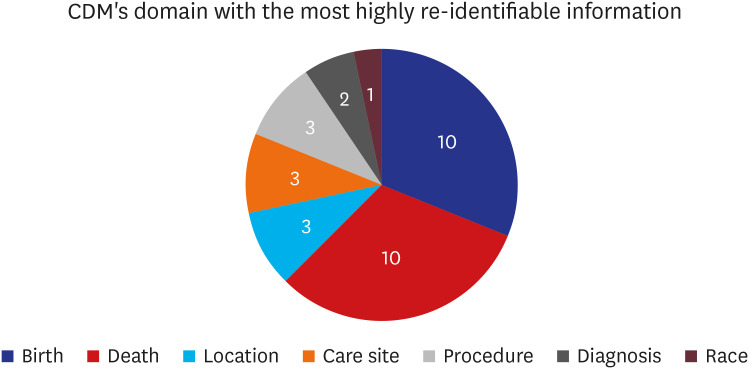
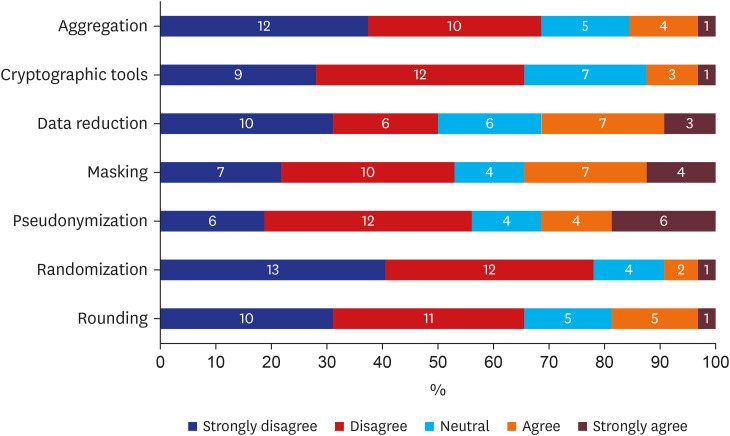
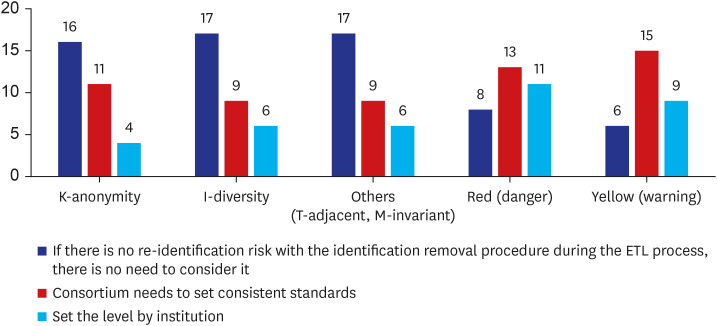
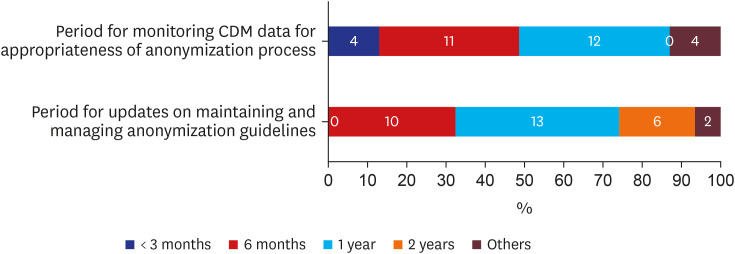
There were 33 valid respondents. Of these, 43.8% suggested additional anonymization was unnecessary beyond, “minimum cell count” policy, which obscures a cell with a value lower than certain number (usually 5) in shared results to minimize the liability of re-identification due to rare conditions. During extract-transform-load processes, 81.8% of respondents assumed structured data is under control from the risk of re-identification. However, respondents noted that date of birth and death were highly re-identifiable information. The majority of respondents (n = 22, 66.7%) conceded the possibility of identifier-contained unstructured data in the NOTE table.
Conclusion
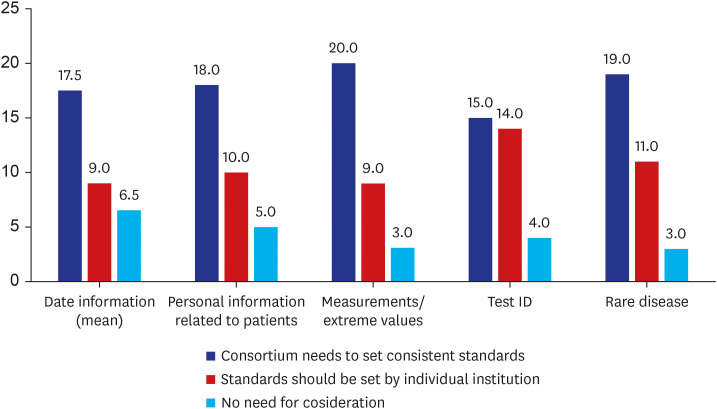
Overall, CDM users generally attributed high reliability for privacy protection to the intrinsic nature of CDM. There was little demand for additional de-identification methods. However, unstructured data in the CDM were suspected to have risks. The necessity for a coordinating consortium to define and manage the re-identification risk of CDM was urged.
Keyword
Figure
Reference
-
1. Benson K, Hartz AJ. A comparison of observational studies and randomized, controlled trials. N Engl J Med. 2000; 342(25):1878–1886. PMID: 10861324.
Article2. Keshta I, Odeh A. Security and privacy of electronic health records: concerns and challenges. Egypt Inform J. 2021; 22(2):177–183.
Article3. Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012; 19(1):54–60. PMID: 22037893.
Article4. You SC, Rho Y, Bikdeli B, Kim J, Siapos A, Weaver J, et al. Association of ticagrelor vs clopidogrel with net adverse clinical events in patients with acute coronary syndrome undergoing percutaneous coronary intervention. JAMA. 2020; 324(16):1640–1650. PMID: 33107944.
Article5. Malenfant JM, Hochstadt J, Nolan B, Barrett K, Corriveau D, Dee D, et al. Cross-Network Directory Service: Infrastructure to enable collaborations across distributed research networks. Learn Health Syst. 2019; 3(2):e10187. PMID: 31245605.
Article6. Jeon S, Seo J, Kim S, Lee J, Kim JH, Sohn JW, et al. Proposal and assessment of a de-identification strategy to enhance anonymity of the observational medical outcomes partnership common data model (OMOP-CDM) in a public cloud-computing environment: anonymization of medical data using privacy models. J Med Internet Res. 2020; 22(11):e19597. PMID: 33177037.
Article7. International Organization for Standardization. Privacy Enhancing Data De-identification Terminology and Classification of Techniques. Geneva, Switzerland: International Organization for Standardization;2018.8. International Organization for Standardization. Health Informatics — Pseudonymization. Geneva, Switzerland: International Organization for Standardization;2017.9. O’Keefe CM, Rubin DB. Individual privacy versus public good: protecting confidentiality in health research. Stat Med. 2015; 34(23):3081–3103. PMID: 26045214.
Article10. Observational Health Data Sciences and Informatics. ATLAS – a unified interface for the OHDSI tools. Accessed May 2, 2022. https://www.ohdsi.org/atlas-a-unified-interface-for-the-ohdsi-tools/ .11. Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015; 216:574–578. PMID: 26262116.12. Khare R, Utidjian LH, Razzaghi H, Soucek V, Burrows E, Eckrich D, et al. Design and refinement of a data quality assessment workflow for a large pediatric research network. EGEMS (Wash DC). 2019; 7(1):36. PMID: 31531382.
Article13. Pfaff ER, Haendel MA, Kostka K, Lee A, Niehaus E, Palchuk MB, et al. Ensuring a safe(r) harbor: excising personally identifiable information from structured electronic health record data. J Clin Transl Sci. 2021; 6(1):e10. PMID: 35211336.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Applying the OMOP Common Data Model to Facilitate Benefit-Risk Assessments of Medicinal Products Using Real-World Data from Singapore and South Korea
- Development and Validation of the Radiology Common Data Model (R-CDM) for the International Standardization of Medical Imaging Data
- Analysis of vancomycin-associated adverse reactions using a common data model
- Utility of Treatment Pattern Analysis Using a Common Data Model: A Scoping Review
- Feasibility Study of Federated Learning on the Distributed Research Network of OMOP Common Data Model