Cancer Res Treat.
2022 Apr;54(2):517-524. 10.4143/crt.2021.206.
Machine Learning Model for Predicting Postoperative Survival of Patients with Colorectal Cancer
- Affiliations
-
- 1Faculty of Medicine, Zagazig University, Zagazig, Egypt
- 2Faculty of Pharmacy, British University in Egypt (BUE), El Shorouk, Egypt
- 3Department of Surgery, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 4Department of Surgery, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2528221
- DOI: http://doi.org/10.4143/crt.2021.206
Abstract
- Purpose
Machine learning (ML) is a strong candidate for making accurate predictions, as we can use large amount of data with powerful computational algorithms. We developed a ML based model to predict survival of patients with colorectal cancer (CRC) using data from two independent datasets.
Materials and Methods
A total of 364,316 and 1,572 CRC patients were included from the Surveillance, Epidemiology, and End Results (SEER) and a Korean dataset, respectively. As SEER combines data from 18 cancer registries, internal validation was done using 18-Fold-Cross-Validation then external validation was performed by testing the trained model on the Korean dataset. Performance was evaluated using area under the receiver operating characteristic curve (AUROC), sensitivity and positive predictive values.
Results
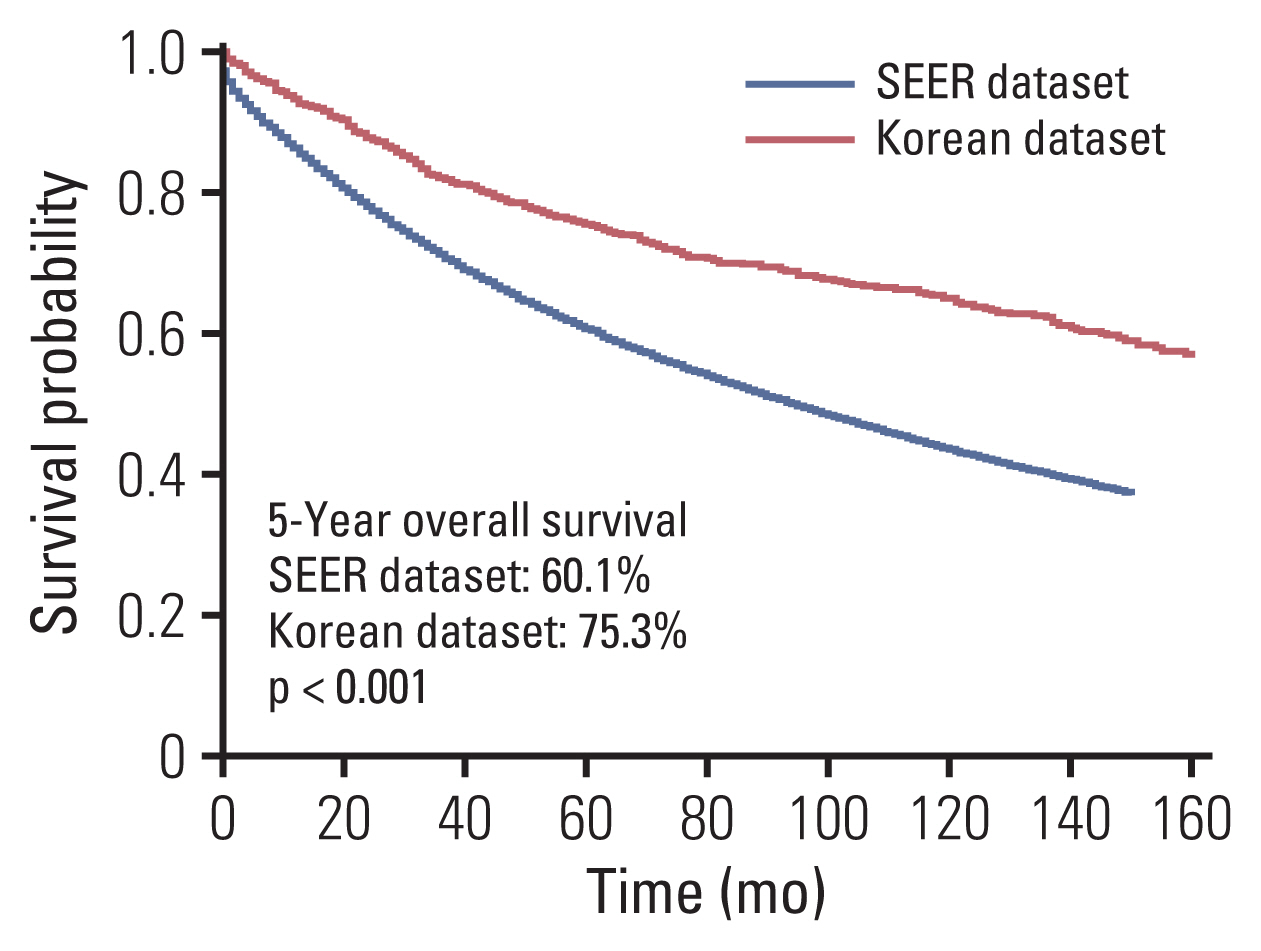
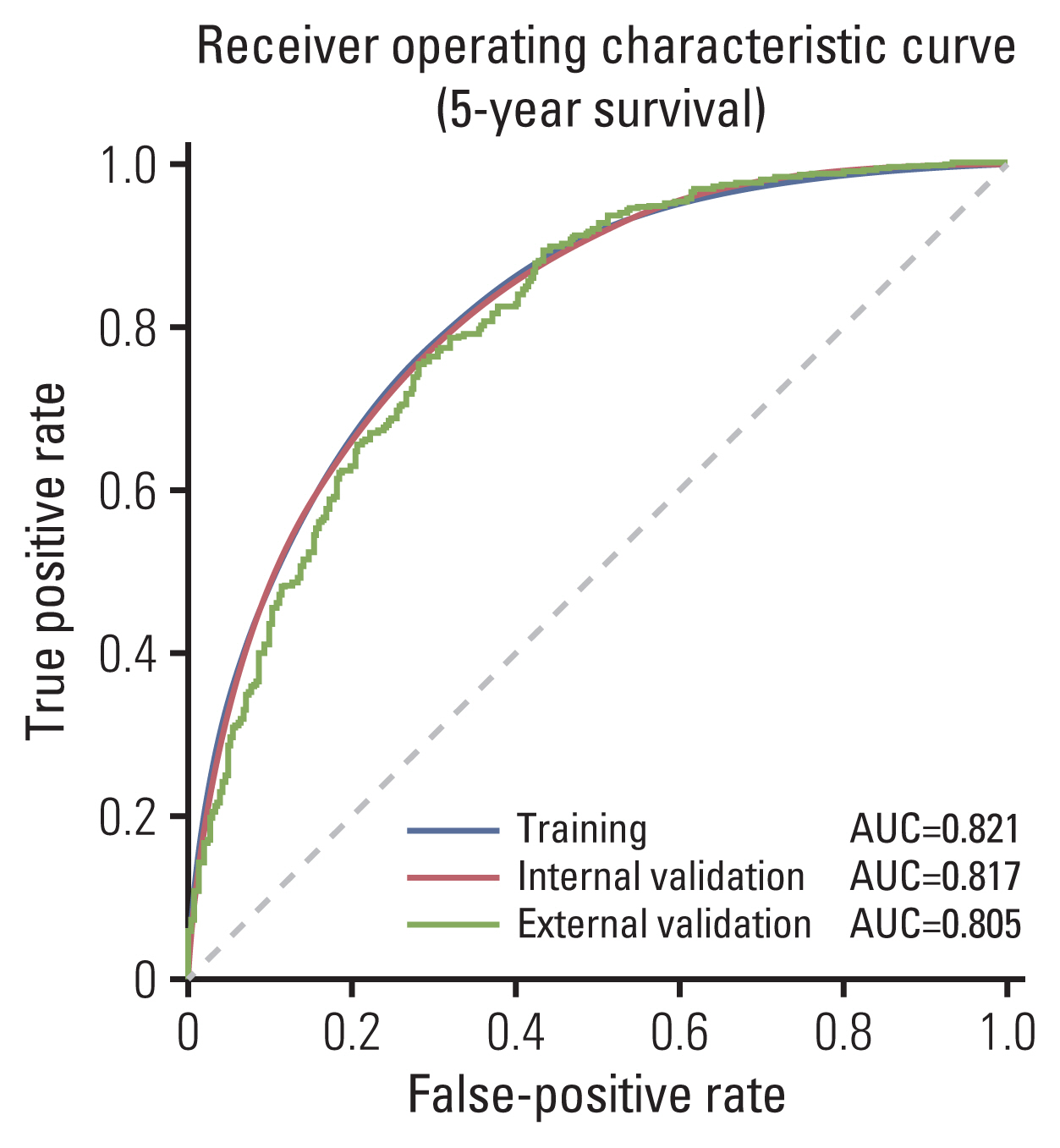
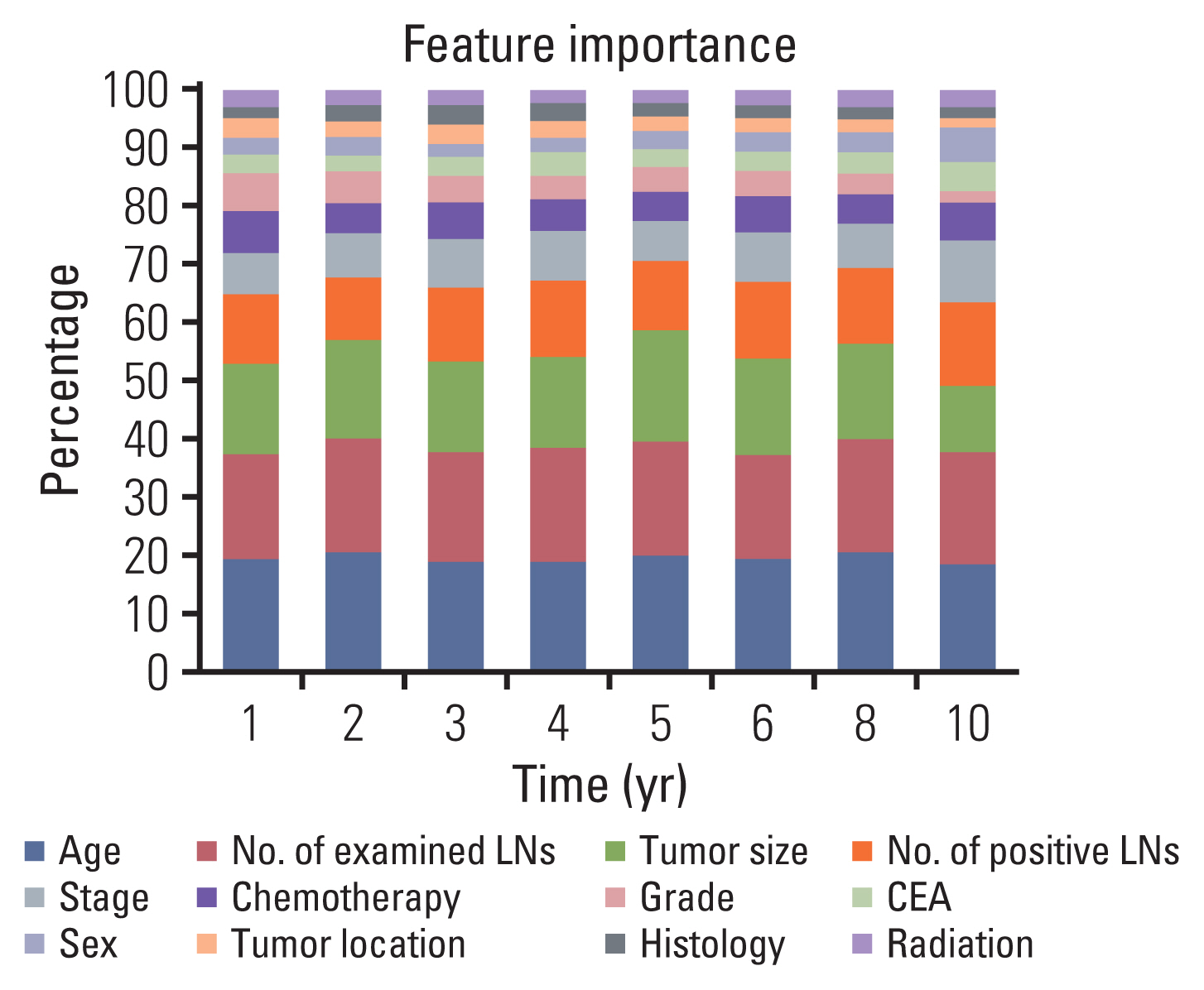
Clinicopathological characteristics were significantly different between the two datasets and the SEER showed a significant lower 5-year survival rate compared to the Korean dataset (60.1% vs. 75.3%, p < 0.001). The ML-based model using the Light gradient boosting algorithm achieved a better performance in predicting 5-year-survival compared to American Joint Committee on Cancer stage (AUROC, 0.804 vs. 0.736; p < 0.001). The most important features which influenced model performance were age, number of examined lymph nodes, and tumor size. Sensitivity and positive predictive values of predicting 5-year-survival for classes including dead or alive were reported as 68.14%, 77.51% and 49.88%, 88.1% respectively in the validation set. Survival probability can be checked using the web-based survival predictor (http://colorectalcancer.pythonanywhere.com).
Conclusion
ML-based model achieved a much better performance compared to staging in individualized estimation of survival of patients with CRC.
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34.
Article2. Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019; 51:417–30.
Article3. NCCN guidelines [Internet]. Plymouth Meeting, PA: National Comprehensive Cancer Network;2019. [cited 2020 Mar 3]. Available from: https://www.nccn.org/professionals/physician_gls/default.aspx .4. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010; 17:1471–4.
Article5. Li J, Guo BC, Sun LR, Wang JW, Fu XH, Zhang SZ, et al. TNM staging of colorectal cancer should be reconsidered by T stage weighting. World J Gastroenterol. 2014; 20:5104–12.
Article6. Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestol K, Maddison J, et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020; 395:350–60.
Article7. Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013; 14:1295–306.
Article8. Haruki K, Kosumi K, Li P, Arima K, Vayrynen JP, Lau MC, et al. An integrated analysis of lymphocytic reaction, tumour molecular characteristics and patient survival in colorectal cancer. Br J Cancer. 2020; 122:1367–77.
Article9. Dercle L, Lu L, Schwartz LH, Qian M, Tejpar S, Eggleton P, et al. Radiomics response signature for identification of metastatic colorectal cancer sensitive to therapies targeting EGFR pathway. J Natl Cancer Inst. 2020; 112:902–12.
Article10. Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smail-Tabbone M, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology. 2020; 158:76–94.
Article11. Reichling C, Taieb J, Derangere V, Klopfenstein Q, Le Malicot K, Gornet JM, et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020; 69:681–90.
Article12. Stojadinovic A, Bilchik A, Smith D, Eberhardt JS, Ward EB, Nissan A, et al. Clinical decision support and individualized prediction of survival in colon cancer: Bayesian belief network model. Ann Surg Oncol. 2013; 20:161–74.
Article13. SEER Cancer Statistics Review, 1975–2016 [Internet]. Bethesda, MD: National Cancer Institute;2019. [cited 2021 Jul 8]. Available from: https://seer.cancer.gov/csr/1975_2016/ .14. Deist TM, Dankers F, Valdes G, Wijsman R, Hsu IC, Oberije C, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: an empirical comparison of classifiers. Med Phys. 2018; 45:3449–59.
Article15. Czarnecki WM, Podlewska S, Bojarski AJ. Robust optimization of SVM hyperparameters in the classification of bioactive compounds. J Cheminform. 2015; 7:38.
Article16. Steyerberg EW, Harrell FE Jr. Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol. 2016; 69:245–7.
Article17. Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982; 143:29–36.
Article18. Elsebaie MA, Amgad M, Elkashash A, Elgebaly AS, Ashal G, Shash E, et al. Management of low and intermediate risk adult rhabdomyosarcoma: a pooled survival analysis of 553 patients. Sci Rep. 2018; 8:9337.
Article19. Lei L, Wang Y, Xue Q, Tong J, Zhou CM, Yang JJ. A comparative study of machine learning algorithms for predicting acute kidney injury after liver cancer resection. PeerJ. 2020; 8:e8583.
Article20. Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020; 70:145–64.
Article21. Schwender H. Imputing missing genotypes with weighted k nearest neighbors. J Toxicol Environ Health A. 2012; 75:438–46.22. Chang GJ, Rodriguez-Bigas MA, Skibber JM, Moyer VA. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2007; 99:433–41.
Article23. Kelder W, Inberg B, Schaapveld M, Karrenbeld A, Grond J, Wiggers T, et al. Impact of the number of histologically exa-mined lymph nodes on prognosis in colon cancer: a population-based study in the Netherlands. Dis Colon Rectum. 2009; 52:260–7.24. Dai W, Mo S, Xiang W, Han L, Li Q, Wang R, et al. The critical role of tumor size in predicting prognosis for T1 colon cancer. Oncologist. 2020; 25:244–51.
Article25. Cha YJ, Park EJ, Baik SH, Lee KY, Kang J. Clinical significance of tumor-infiltrating lymphocytes and neutrophil-to-lymphocyte ratio in patients with stage III colon cancer who underwent surgery followed by FOLFOX chemotherapy. Sci Rep. 2019; 9:11617.
Article26. Kang J, Kim H, Hur H, Min BS, Baik SH, Lee KY, et al. Circumferential resection margin involvement in stage III rectal cancer patients treated with curative resection followed by chemoradiotherapy: a surrogate marker for local recurrence? Yonsei Med J. 2013; 54:131–8.
Article27. Hwang MR, Park JW, Park S, Yoon H, Kim DY, Chang HJ, et al. Prognostic impact of circumferential resection margin in rectal cancer treated with preoperative chemoradiotherapy. Ann Surg Oncol. 2014; 21:1345–51.
Article28. Lee JM, Chung T, Kim KM, Simon NS, Han YD, Cho MS, et al. Significance of radial margin in patients undergoing complete mesocolic excision for colon cancer. Dis Colon Rectum. 2020; 63:488–96.
Article29. Badic B, Hatt M, Durand S, Jossic-Corcos CL, Simon B, Visvikis D, et al. Radiogenomics-based cancer prognosis in colorectal cancer. Sci Rep. 2019; 9:9743.
Article30. Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017; 17:79–92.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning-Based Predictor for Treatment Outcomes of Patients With Salivary Gland Cancer After Operation
- Radiomics and machine learning analysis of liver magnetic resonance imaging for prediction and early detection of tumor response in colorectal liver metastases
- Clinico-pathologic Factors and Machine Learning Algorithm for Survival Prediction in Parotid Gland Cancer
- Machine Learning Algorithms for Predicting Treatment Outcomes of Oropharyngeal Cancer After Surgery
- Diagnostic Accuracy of Machine Learning Algorithms for Hepatitis A Antibody