J Korean Med Sci.
2022 Feb;37(5):e33. 10.3346/jkms.2022.37.e33.
Flat Pattern Peaks of Tacrolimus Absorption and Associated Pharmacogenomic Variants in Kidney Transplantation Recipients
- Affiliations
-
- 1Department of Surgery, Chung-Ang University College of Medicine, Seoul, Korea
- 2Seoul National University Biomedical Informatics (SNUBI), Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Surgery, Seoul National University College of Medicine, Seoul, Korea
- 4Department of Information Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- 5Transplantation Research Institute, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2525485
- DOI: http://doi.org/10.3346/jkms.2022.37.e33
Abstract
- Background
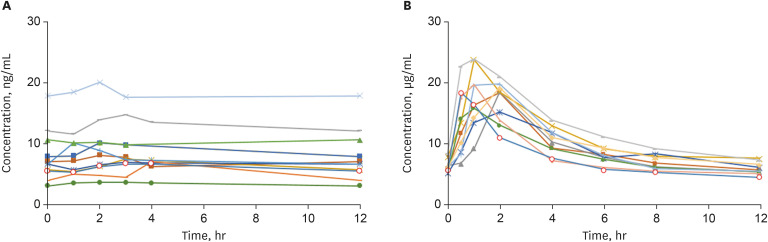
Tacrolimus is the most commonly used immunosuppressive drug in solid organ transplantation. After administering a conventional twice-daily dose of tacrolimus, peak levels were achieved within the first 1.5 to 2 hours. A group of patients showed different early absorption phase of tacrolimus after kidney transplantation.
Methods
Trough(C0 ) and 1.5-hour blood levels (C1.5 ) of tacrolimus were measured in 95 kidney transplantation recipients. Patients with a C1.5 /C0 < 1.5 and > 1.5 were defined as those having flat pattern peaks and as controls, respectively. Transplantation outcomes were compared between the groups. Whole exome sequencing was performed to investigate the genetic susceptibility to flat pattern peaks.
Results
Twenty-eight patients showed flat pattern peaks. The mean C1.5 /C0 values were 1.13 ± 0.22 and 3.78 ± 1.25 in the flat pattern peak and control groups, respectively. In multivariate analysis, flat pattern peak was an independent risk factor for biopsy-proven acute rejection (BPAR) and/or borderline change (P = 0.014). Patients having flat pattern peaks showed significantly lower post-transplant 36-month estimated glomerular filtration rate (P = 0.001). Two single nucleotide variants in ABCB1 genes, rs1922242 and rs2235035, were associated with flat pattern peaks (P = 0.019 and P = 0.027, respectively).
Conclusion
Both of C1.5 and C0should be measured to distinguish the patients showing unique initial absorption. A C1.5 /C0 ratio lower than 1.5 was associated with an increased risk of BPAR and/or borderline change. Single nucleotide variants s in ABCB1 gene might influence the flat pattern peaks of tacrolimus absorption.
Keyword
Figure
Reference
-
1. Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015; 98(1):19–24. PMID: 25801146.2. Schiff J, Cole E, Cantarovich M. Therapeutic monitoring of calcineurin inhibitors for the nephrologist. Clin J Am Soc Nephrol. 2007; 2(2):374–384. PMID: 17699437.
Article3. Stifft F, Stolk LM, Undre N, van Hooff JP, Christiaans MH. Lower variability in 24-hour exposure during once-daily compared to twice-daily tacrolimus formulation in kidney transplantation. Transplantation. 2014; 97(7):775–780. PMID: 24686426.
Article4. Undre NA. Pharmacokinetics of tacrolimus-based combination therapies. Nephrol Dial Transplant. 2003; 18(Suppl 1):i12–i15. PMID: 12738758.
Article5. Mathew BS, Fleming DH, Jeyaseelan V, Chandy SJ, Annapandian VM, Subbanna PK, et al. A limited sampling strategy for tacrolimus in renal transplant patients. Br J Clin Pharmacol. 2008; 66(4):467–472. PMID: 18662286.
Article6. Picard N, Bergan S, Marquet P, van Gelder T, Wallemacq P, Hesselink DA, et al. Pharmacogenetic biomarkers predictive of the pharmacokinetics and pharmacodynamics of immunosuppressive drugs. Ther Drug Monit. 2016; 38(Suppl 1):S57–S69. PMID: 26469711.
Article7. Birdwell K. Role of pharmacogenomics in dialysis and transplantation. Curr Opin Nephrol Hypertens. 2014; 23(6):570–577. PMID: 25162201.
Article8. Sohn M, Kim MG, Han N, Kim IW, Gim J, Min SI, et al. Whole exome sequencing for the identification of CYP3A7 variants associated with tacrolimus concentrations in kidney transplant patients. Sci Rep. 2018; 8(1):18064. PMID: 30584253.
Article9. Roufosse C, Simmonds N, Clahsen-van Groningen M, Haas M, Henriksen KJ, Horsfield C, et al. A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation. 2018; 102(11):1795–1814. PMID: 30028786.
Article10. Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al. The Banff 2015 Kidney Meeting Report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017; 17(1):28–41. PMID: 27862883.
Article11. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43(5):491–498. PMID: 21478889.
Article12. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38(16):e164. PMID: 20601685.
Article13. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011; 29(1):24–26. PMID: 21221095.
Article14. Trofe-Clark J, Lemonovich TL. AST Infectious Diseases Community of Practice. Interactions between anti-infective agents and immunosuppressants in solid organ transplantation. Am J Transplant. 2013; 13(Suppl 4):318–326. PMID: 23465024.
Article15. U.S. Food & Drug Administration. Updated 2020. Accessed June 14, 2021. https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers .16. Clayton PA, McDonald SP, Russ GR, Chadban SJ. Long-term outcomes after acute rejection in kidney transplant recipients: an ANZDATA analysis. J Am Soc Nephrol. 2019; 30(9):1697–1707. PMID: 31308074.
Article17. Seifert ME, Agarwal G, Bernard M, Kasik E, Raza SS, Fatima H, et al. Impact of subclinical borderline inflammation on kidney transplant outcomes. Transplant Direct. 2021; 7(2):e663. PMID: 33511268.
Article18. Min SI, Park YS, Ahn S, Park T, Park DD, Kim SM, et al. Chronic allograft injury by subclinical borderline change: evidence from serial protocol biopsies in kidney transplantation. J Korean Surg Soc. 2012; 83(6):343–351. PMID: 23230552.
Article19. Aouam K, Chadli Z, Hammouda M, Fredj NB, Aloui S, May ME, et al. Development of limited sampling strategies for the estimation of tacrolimus area under the curve in adult kidney transplant recipients according to the posttransplantation time. Ther Drug Monit. 2015; 37(4):524–530. PMID: 25627405.
Article20. Nakazawa R, Yoshiike M, Nozawa S, Aida K, Katsuoka Y, Fujimoto E, et al. Clinically useful limited sampling strategy to estimate area under the concentration-time curve of once-daily tacrolimus in adult Japanese kidney transplant recipients. PLoS One. 2019; 14(12):e0225878. PMID: 31825991.
Article21. Brunet M, van Gelder T, Åsberg A, Haufroid V, Hesselink DA, Langman L, et al. Therapeutic drug monitoring of tacrolimus-personalized therapy: second consensus report. Ther Drug Monit. 2019; 41(3):261–307. PMID: 31045868.
Article22. Khan AR, Raza A, Firasat S, Abid A. CYP3A5 gene polymorphisms and their impact on dosage and trough concentration of tacrolimus among kidney transplant patients: a systematic review and meta-analysis. Pharmacogenomics J. 2020; 20(4):553–562. PMID: 31902947.
Article23. Huang L, Wang J, Yang J, Zhang H, Ni Y, Zhu Z, et al. Impact of CYP3A4/5 and ABCB1 polymorphisms on tacrolimus exposure and response in pediatric primary nephrotic syndrome. Pharmacogenomics. 2019; 20(15):1071–1083. PMID: 31588879.24. Genvigir FD, Campos-Salazar AB, Felipe CR, Tedesco-Silva H Jr, Medina-Pestana JO, Doi SQ, et al. CYP3A5*3 and CYP2C8*3 variants influence exposure and clinical outcomes of tacrolimus-based therapy. Pharmacogenomics. 2020; 21(1):7–21. PMID: 31849280.25. Staatz CE, Goodman LK, Tett SE. Effect of CYP3A and ABCB1 single nucleotide polymorphisms on the pharmacokinetics and pharmacodynamics of calcineurin inhibitors: part I. Clin Pharmacokinet. 2010; 49(3):141–175. PMID: 20170205.
Article26. Zhang Y, Benet LZ. The gut as a barrier to drug absorption: combined role of cytochrome P450 3A and P-glycoprotein. Clin Pharmacokinet. 2001; 40(3):159–168. PMID: 11327196.27. Hesselink DA, Bouamar R, Elens L, van Schaik RH, van Gelder T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin Pharmacokinet. 2014; 53(2):123–139. PMID: 24249597.
Article28. Vanhove T, Annaert P, Lambrechts D, Kuypers DR. Effect of ABCB1 diplotype on tacrolimus disposition in renal recipients depends on CYP3A5 and CYP3A4 genotype. Pharmacogenomics J. 2017; 17(6):556–562. PMID: 27378609.
Article29. Min SI, Ha J, Kim YS, Ahn SH, Park T, Park DD, et al. Therapeutic equivalence and pharmacokinetics of generic tacrolimus formulation in de novo kidney transplant patients. Nephrol Dial Transplant. 2013; 28(12):3110–3119. PMID: 24084327.
Article30. Tremblay S, Nigro V, Weinberg J, Woodle ES, Alloway RR. A steady-state head-to-head pharmacokinetic comparison of all FK-506 (Tacrolimus) formulations (ASTCOFF): an open-label, prospective, randomized, two-arm, three-period crossover study. Am J Transplant. 2017; 17(2):432–442. PMID: 27340950.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case Report of Kidney Graft Failure after Duodenal Bypass Surgery
- Modified Release Tacrolimus
- Comparison of Tacrolimus and Cyclosporine in Kidney Transplantation
- Risk of graft loss on once-daily versus twice-daily tacrolimus in kidney transplant patients: a meta-analysis
- Severe anorexia and weight loss induced by tacrolimus in kidney transplant recipients