Cancer Res Treat.
2021 Jul;53(3):754-762. 10.4143/crt.2020.478.
Association between ALDH2 and ADH1B Polymorphisms and the Risk for Colorectal Cancer in Koreans
- Affiliations
-
- 1Department of Preventive Medicine, Chonnam National University Medical School, Hwasun, Korea
- 2Department of Hematology-Oncology, Chonnam National University Hwasun Hospital, Hwasun, Korea
- 3Gwangju-Jeonnam Regional Cardiocerebrovascular Center, Chonnam National University Hospital, Gwangju, Korea
- 4Vanderbilt-Ingram Cancer Center, Vanderbilt Epidemiology Center, Vanderbilt University School of Medicine, Nashville, TN, USA
- KMID: 2518399
- DOI: http://doi.org/10.4143/crt.2020.478
Abstract
- Purpose
Excessive alcohol consumption has been linked to an increased risk of colorectal cancer (CRC). We evaluated the association between alcohol-related genetic variants and CRC risk.
Materials and Methods
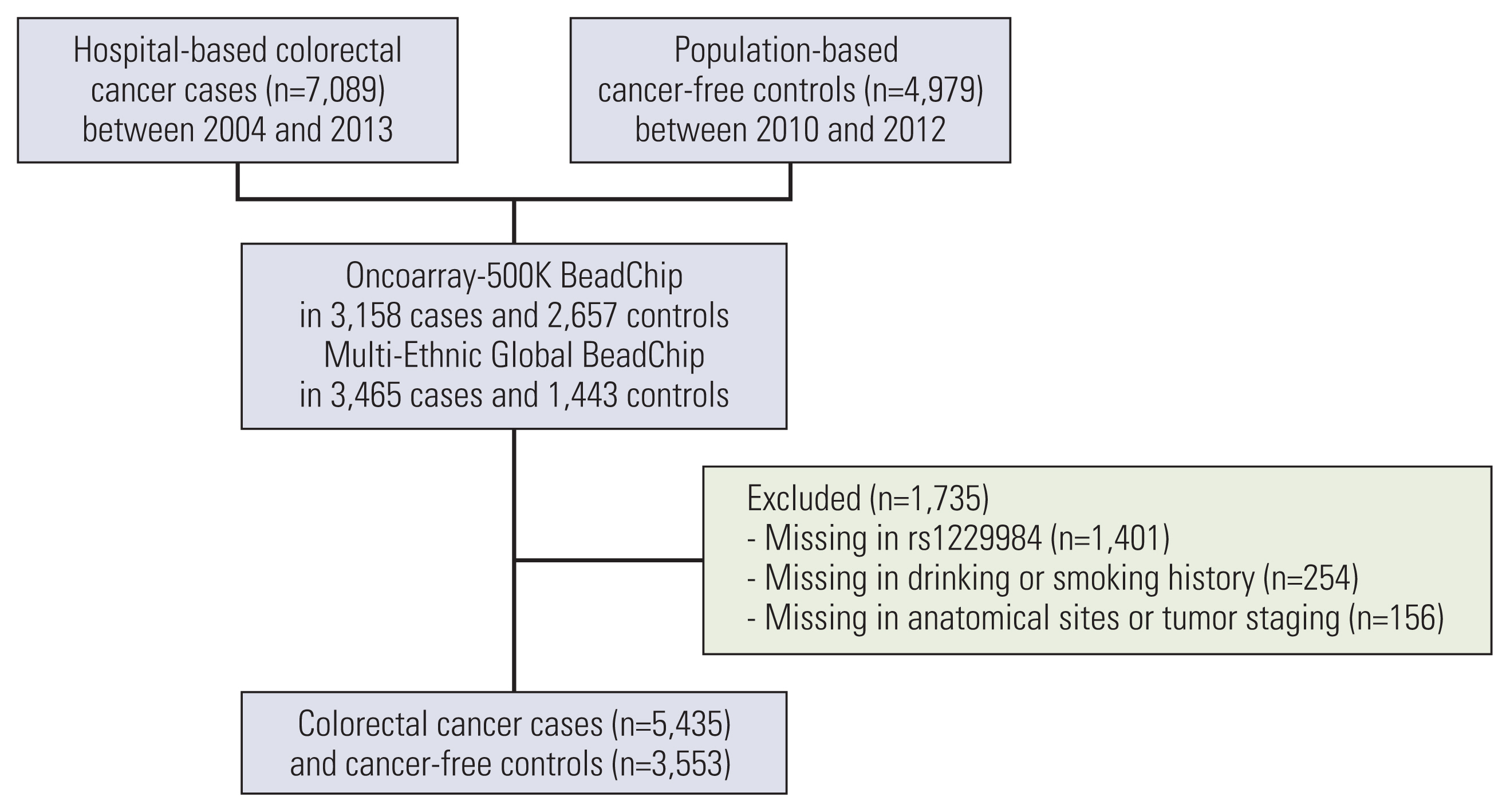
The study cohort consisted of 5,435 CRC cases and 3,553 population-based cancer-free controls. Genotype data were generated from germline DNA using the Infinium OncoArray-500K BeadChip in 2,535 cases and 2,287 controls and the Infinium Multi-Ethnic Global BeadChip in 2,900 cases and 1,266 controls. The associations between aldehyde dehydrogenase 2 (ALDH2) rs671 and alcohol dehydrogenase 1B (ADH1B) rs1229984 polymorphisms and CRC risk were assessed using multivariate logistic regression analyses.
Results
Compared with the major homozygous ALDH2 genotype (GG), heterozygous or minor homozygous ALDH2 genotype (GA or AA, related to a low alcohol consumption) was significantly associated with a reduced risk for CRC in men (odds ratio [OR], 0.78; 95% confidence interval [CI], 0.68 to 0.90), but not in women (OR, 0.70; 95% CI, 0.47 to 1.05). A stronger association was found among regular drinkers (OR, 0.58; 95% CI, 0.47 to 0.71 in men and OR, 0.33; 95% CI, 0.18 to 0.58 in women). No association of CRC risk with ADH1B rs1229984 genotype was found. The association between alcohol-related combined genotypes and risk of CRC was significant (p for linear=0.001). The combined genotype with the highest genetically predicted alcohol consumption (ALDH2 rs671 GG and ADH1B rs1229984 AG/GG) was associated with a high risk for CRC (OR, 1.35; 95% CI, 1.11 to 1.63).
Conclusion
Our study provides strong evidence for a possible causal association between alcohol consumption and CRC risk.
Figure
Reference
-
References
1. Kweon SS. Updates on cancer epidemiology in Korea, 2018. Chonnam Med J. 2018; 54:90–100.
Article2. Cai S, Li Y, Ding Y, Chen K, Jin M. Alcohol drinking and the risk of colorectal cancer death: a meta-analysis. Eur J Cancer Prev. 2014; 23:532–9.3. GBD 2016 Alcohol Collaborators. Alcohol use and burden for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2018; 392:1015–35.4. Klarich DS, Brasser SM, Hong MY. Moderate alcohol consumption and colorectal cancer risk. Alcohol Clin Exp Res. 2015; 39:1280–91.
Article5. Lee S, Woo H, Lee J, Oh JH, Kim J, Shin A. Cigarette smoking, alcohol consumption, and risk of colorectal cancer in South Korea: a case-control study. Alcohol. 2019; 76:15–21.
Article6. McNabb S, Harrison TA, Albanes D, Berndt SI, Brenner H, Caan BJ, et al. Meta-analysis of 16 studies of the association of alcohol with colorectal cancer. Int J Cancer. 2020; 146:861–73.
Article7. Cornish AJ, Tomlinson IPM, Houlston RS. Mendelian randomisation: a powerful and inexpensive method for identifying and excluding non-genetic risk factors for colorectal cancer. Mol Aspects Med. 2019; 69:41–7.
Article8. Millwood IY, Walters RG, Mei XW, Guo Y, Yang L, Bian Z, et al. Conventional and genetic evidence on alcohol and vascular disease aetiology: a prospective study of 500 000 men and women in China. Lancet. 2019; 393:1831–42.9. Kim DJ, Choi IG, Park BL, Lee BC, Ham BJ, Yoon S, et al. Major genetic components underlying alcoholism in Korean population. Hum Mol Genet. 2008; 17:854–8.
Article10. Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, et al. Confirmation of ALDH2 as a major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011; 75:911–8.
Article11. Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014; 165B:103–10.12. Cho Y, Kwak S, Lewis SJ, Wade KH, Relton CL, Smith GD, et al. Exploring the utility of alcohol flushing as an instrumental variable for alcohol intake in Koreans. Sci Rep. 2018; 8:458.
Article13. Yin G, Kono S, Toyomura K, Moore MA, Nagano J, Mizoue T, et al. Alcohol dehydrogenase and aldehyde dehydrogenase polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 2007; 98:1248–53.
Article14. Chiang CP, Jao SW, Lee SP, Chen PC, Chung CC, Lee SL, et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human large bowel: association of the functional polymorphisms of ADH and ALDH genes with hemorrhoids and colorectal cancer. Alcohol. 2012; 46:37–49.
Article15. Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Suzuki T, et al. A gene-gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis. 2006; 27:1018–23.
Article16. Yang H, Zhou Y, Zhou Z, Liu J, Yuan X, Matsuo K, et al. A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev. 2009; 18:2522–7.
Article17. Ferrari P, McKay JD, Jenab M, Brennan P, Canzian F, Vogel U, et al. Alcohol dehydrogenase and aldehyde dehydrogenase gene polymorphisms, alcohol intake and the risk of colorectal cancer in the European Prospective Investigation into Cancer and Nutrition study. Eur J Clin Nutr. 2012; 66:1303–8.
Article18. Zhao H, Liu KJ, Lei ZD, Lei SL, Tian YQ. Meta-analysis of the aldehyde dehydrogenases-2 (ALDH2) Glu487Lys polymorphism and colorectal cancer risk. PLoS One. 2014; 9:e88656.
Article19. Guo XF, Wang J, Yu SJ, Song J, Ji MY, Zhang JX, et al. Meta-analysis of the ADH1B and ALDH2 polymorphisms and the risk of colorectal cancer in East Asians. Intern Med. 2013; 52:2693–9.
Article20. Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014; 46:533–42.
Article21. Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai Q, et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019; 156:1455–66.
Article22. Schmit SL, Edlund CK, Schumacher FR, Gong J, Harrison TA, Huyghe JR, et al. Novel common genetic susceptibility loci for colorectal cancer. J Natl Cancer Inst. 2019; 111:146–57.23. Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015; 526:68–74.24. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012; 44:955–9.
Article25. Coleman JR, Euesden J, Patel H, Folarin AA, Newhouse S, Breen G. Quality control, imputation and analysis of genome-wide genotyping data from the Illumina HumanCoreExome microarray. Brief Funct Genomics. 2016; 15:298–304.
Article26. Matsuo K, Hamajima N, Hirai T, Kato T, Koike K, Inoue M, et al. Aldehyde dehydrogenase 2 (ALDH2) genotype affects rectal cancer susceptibility due to alcohol consumption. J Epidemiol. 2002; 12:70–6.
Article27. Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1-B*2 on blood acetaldehyde concentrations. Hum Genomics. 2009; 3:121–7.
Article28. Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Yokoyama T. Contribution of the alcohol dehydrogenase-1B genotype and oral microorganisms to high salivary acetaldehyde concentrations in Japanese alcoholic men. Int J Cancer. 2007; 121:1047–54.
Article29. Nieminen MT, Salaspuro M. Local acetaldehyde: an essential role in alcohol-related upper gastrointestinal tract carcinogenesis. Cancers (Basel). 2018; 10:11.30. Burgess S, Thompson SG. CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011; 40:755–64.
Article31. Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015; 4:193–9.
Article32. Ugai T, Milne RL, Ito H, Aronson KJ, Bolla MK, Chan T, et al. The functional ALDH2 polymorphism is associated with breast cancer risk: a pooled analysis from the Breast Cancer Association Consortium. Mol Genet Genomic Med. 2019; 7:e707.
Article33. Roerecke M, Tobe SW, Kaczorowski J, Bacon SL, Vafaei A, Hasan OS, et al. Sex-specific associations between alcohol consumption and incidence of hypertension: a systematic review and meta-analysis of cohort studies. J Am Heart Assoc. 2018; 7:e008202.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Influence of ADH1B, ALDH2 Activities and Their Combination on Drinking Behaviors of Korean Young Adults
- The Genetic Factors Affecting Drinking Behaviors of Korean Young Adults with Variant Aldehyde Dehydrogenase 2 Genotype
- Association between ALDH2 Polymorphism and Gastric Cancer Risk in a Korean Population
- A genome-wide association study identified genetic loci for end-stage liver disease in the Korean population
- Association of Dietary Vitamin D and Calcium With Genetic Polymorphisms in Colorectal Neoplasia