Cancer Res Treat.
2021 Apr;53(2):399-408. 10.4143/crt.2020.870.
Upregulated N6-Methyladenosine RNA in Peripheral Blood: Potential Diagnostic Biomarker for Breast Cancer
- Affiliations
-
- 1Medical School, Southeast University, Nanjing, China
- 2Center of Clinical Laboratory Medicine, Zhongda Hospital, Southeast University, Nanjing, China
- KMID: 2514922
- DOI: http://doi.org/10.4143/crt.2020.870
Abstract
- Purpose
An effective biomarker for the diagnosis of breast cancer (BC) and benign breast diseases (BBD) is crucial for improving the prognosis. We investigated whether N6-methyladenosine (m6A) can be a diagnostic biomarker of BC.
Materials and Methods
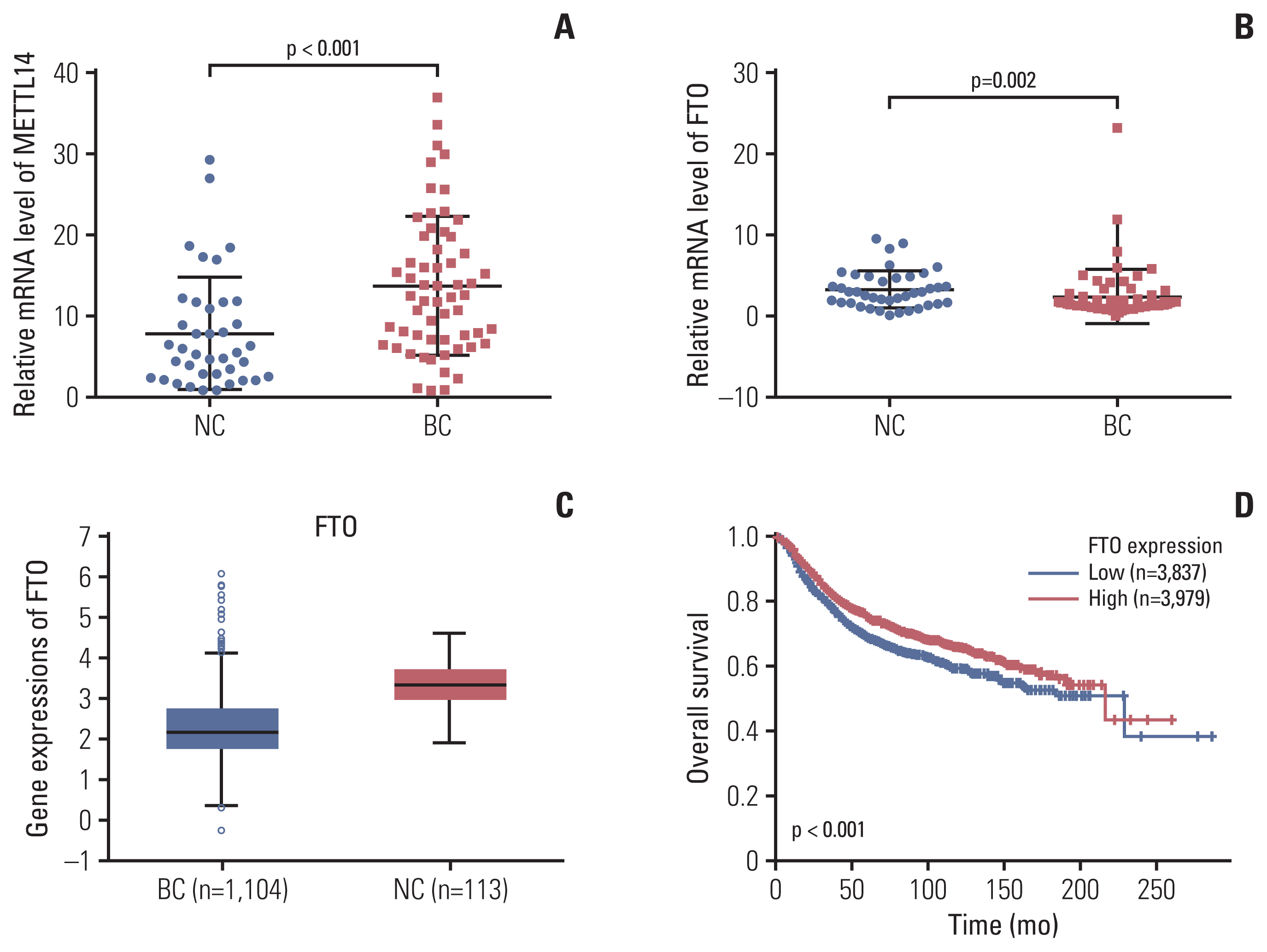
We detected the contents of peripheral blood m6A in 62 patients with BC, 41 patients with BBD, and 41 normal controls (NCs) using the colorimetric method. The relative expression of the m6A regulated genes methyltransferase-like 14 (METTL14) and fat mass and obesity-associated (FTO) was analyzed using quantitative real-time polymerase chain reaction.
Results
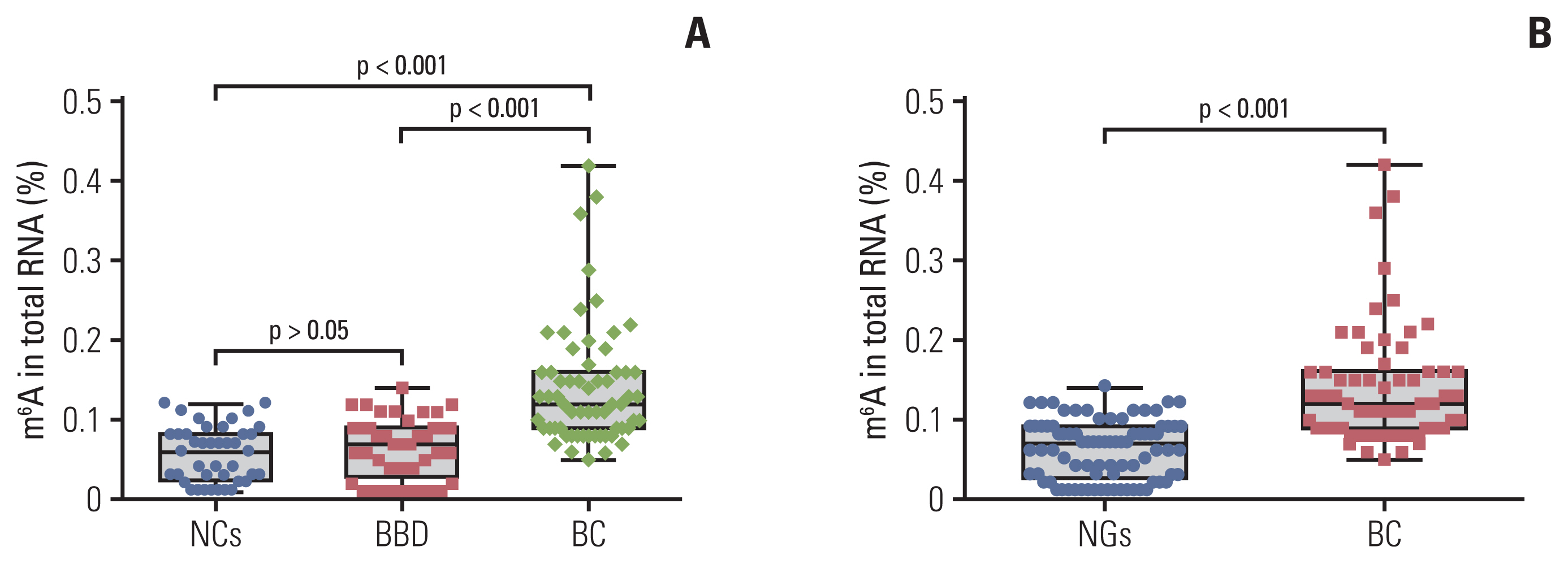
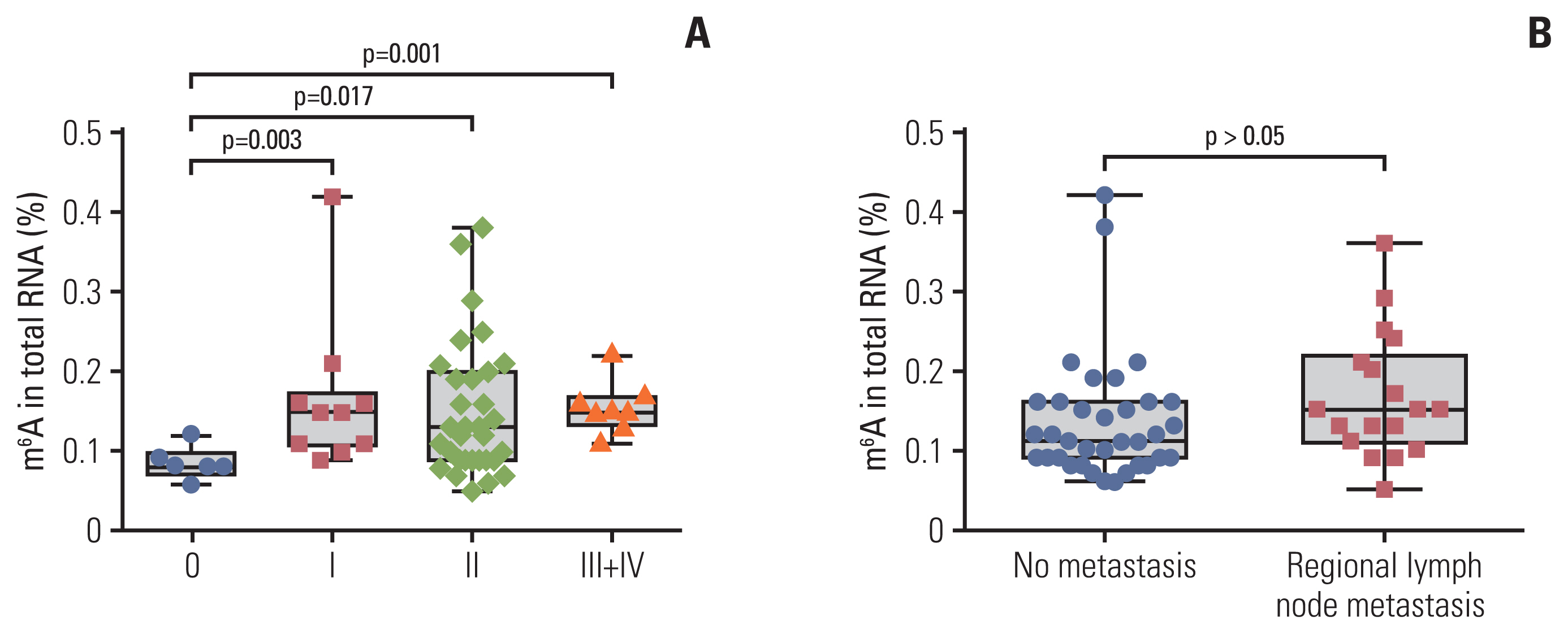
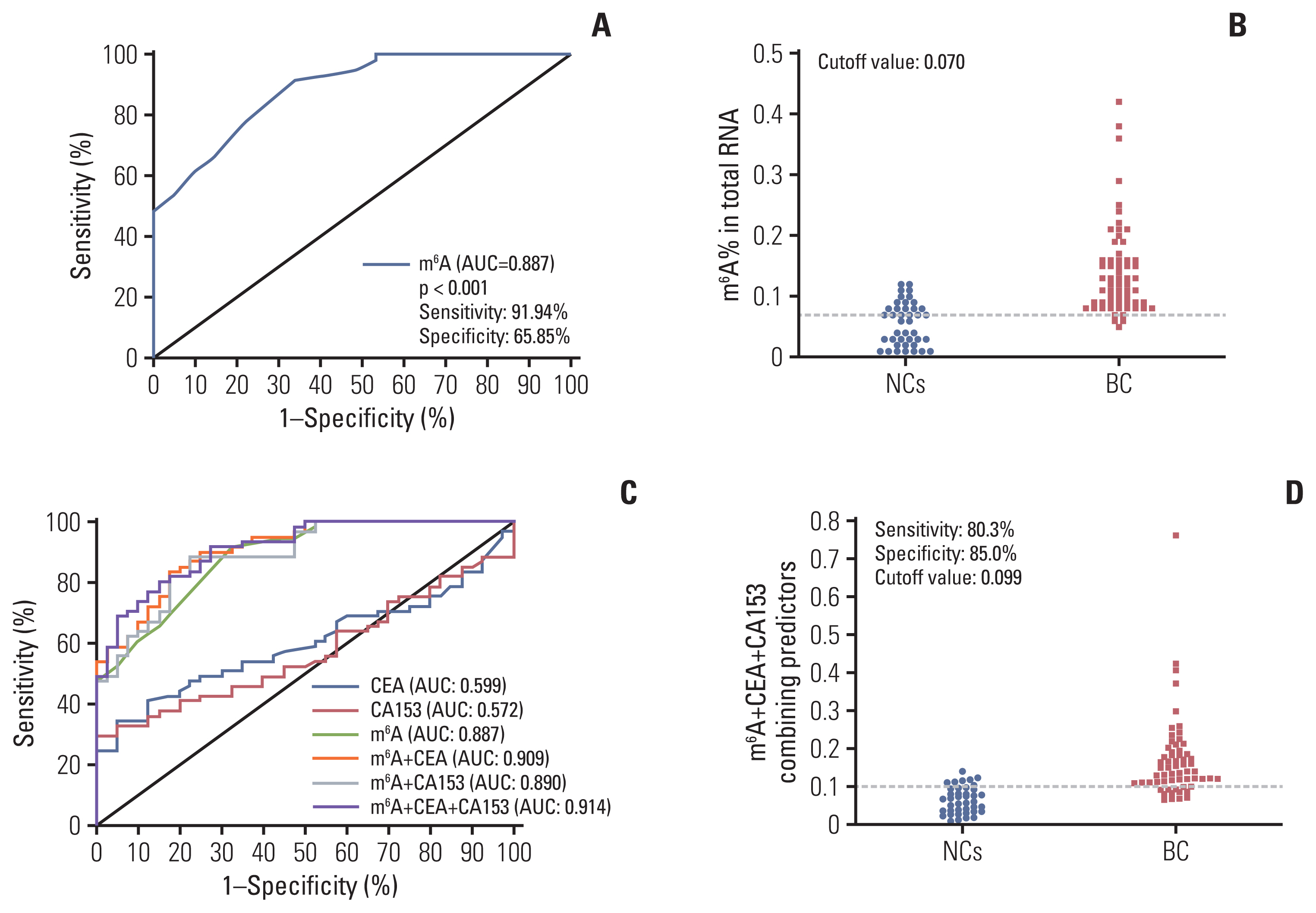
m6A in peripheral blood RNA was significantly higher in patients with BC than that in patients with BBD (p < 0.001) or the NCs (p < 0.001). m6A was closely associated with the disease stage (from stage 0 to stage I-IV, p=0.003). The receiver operating characteristic curve of m6A contained an area under the curve (AUC) value of 0.887 in BC, which was greater than that of carcinoembryonic antigen (CEA) or carbohydrate antigen 153 (CA153). The combination of m6A, CEA, and CA153 improved the AUC to 0.914. The upregulated and downregulated mRNA expression of METTL14 and FTO, respectively, might contribute to the increase of m6A in patients with BC. m6A combined with METTL14 and FTO improved the AUC to 0.929 with a specificity of 97.4% in the peripheral blood of patients with BC.
Conclusion
The peripheral blood RNA of m6A might be a valuable biomarker for the diagnosis of BC.
Keyword
Figure
Reference
-
References
1. Zhang L, Qiang J, Yang X, Wang D, Rehman AU, He X, et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci (Weinh). 2020; 7:1901728.
Article2. Tang J, Ren J, Cui Q, Zhang D, Kong D, Liao X, et al. A prognostic 10-lncRNA expression signature for predicting the risk of tumour recurrence in breast cancer patients. J Cell Mol Med. 2019; 23:6775–84.
Article3. Chavez-MacGregor M, Mittendorf EA, Clarke CA, Lichtensztajn DY, Hunt KK, Giordano SH. Incorporating tumor characteristics to the American Joint Committee on Cancer breast cancer staging system. Oncologist. 2017; 22:1292–300.
Article4. Gao Y, Liu M, Shi S, Sun Y, Li M, Zhang M, et al. Diagnostic value of seven biomarkers for breast cancer: an overview with evidence mapping and indirect comparisons of diagnostic test accuracy. Clin Exp Med. 2020; 20:97–108.
Article5. Nam SE, Lim W, Jeong J, Lee S, Choi J, Park H, et al. The prognostic significance of preoperative tumor marker (CEA, CA15-3) elevation in breast cancer patients: data from the Korean Breast Cancer Society Registry. Breast Cancer Res Treat. 2019; 177:669–78.
Article6. Koh CW, Goh YT, Goh WS. Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat Commun. 2019; 10:5636.
Article7. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019; 18:176.
Article8. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019; 18:178.
Article9. Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019; 18:181.
Article10. Deng X, Su R, Feng X, Wei M, Chen J. Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018; 48:1–7.
Article11. Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016; 113:E2047–56.
Article12. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019; 79:1285–92.
Article13. Cheng J, Tang Q, Cao X, Burwinkel B. Cell-free circulating DNA integrity based on peripheral blood as a biomarker for diagnosis of cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2017; 26:1595–602.
Article14. Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015; 100:E148–54.15. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010; 17:1471–4.
Article16. Pasculli B, Barbano R, Parrella P. Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin Cancer Biol. 2018; 51:22–35.
Article17. Di Gioia D, Blankenburg I, Nagel D, Heinemann V, Stieber P. Tumor markers in the early detection of tumor recurrence in breast cancer patients: CA 125, CYFRA 21-1, HER2 shed antigen, LDH and CRP in combination with CEA and CA 15-3. Clin Chim Acta. 2016; 461:1–7.
Article18. Bodelon C, Ambatipudi S, Dugue PA, Johansson A, Sampson JN, Hicks B, et al. Blood DNA methylation and breast cancer risk: a meta-analysis of four prospective cohort studies. Breast Cancer Res. 2019; 21:62.
Article19. Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenetics. 2016; 8:115.
Article20. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018; 561:556–60.
Article21. Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020; 722:144076.
Article22. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018; 28:507–17.
Article23. Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019; 145:19–29.
Article24. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017; 31:127–41.25. Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019; 10:5447–59.
Article26. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019; 19:326.
Article27. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016; 30:836–48.
Article28. Halvorsen AR, Helland A, Gromov P, Wielenga VT, Talman MM, Brunner N, et al. Profiling of microRNAs in tumor interstitial fluid of breast tumors: a novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol Oncol. 2017; 11:220–34.29. Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis Seeding. Cell. 2019; 176:98–112.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Suppression Effects of Fat Mass and Obesity Associated Gene on the Hair Follicle-Derived Neural Crest Stem Cells Differentiating into Melanocyte by N6-Methyladenosine Modifying Microphthalmia-Associated Transcription Factor
- Potential of the Microbiome as a Biomarker for Early Diagnosis and Prognosis of Breast Cancer
- Potential Biomarker of L-type Amino Acid Transporter 1 in Breast Cancer Progression
- c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer
- N6-Methyladenosine Methyltransferase METTL3 Alleviates Diabetes-Induced Testicular Damage through Modulating TUG1/Clusterin Axis