Cancer Res Treat.
2021 Apr;53(2):389-398. 10.4143/crt.2020.694.
Clinical and Genomic Characteristics of Adult Diffuse Midline Glioma
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea
- 2Cancer Research Institute, Seoul National University, Seoul, Korea
- 3Department of Pathology, Seoul National University Hospital, Seoul, Korea
- 4Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- 5Department of Neurosurgery, Seoul National University Hospital, Seoul, Korea
- 6Department of Radiology, Seoul National University Hospital, Seoul, Korea
- 7Department of Neurology, Seoul National University Hospital, Seoul, Korea
- 8Department of Radiation Oncology, Seoul National University Hospital, Seoul, Korea
- KMID: 2514921
- DOI: http://doi.org/10.4143/crt.2020.694
Abstract
- Purpose
The treatment outcomes and genomic profiles of diffuse midline glioma (DMG) in adult patients are rarely characterized. We performed a retrospective study to evaluate the clinicogenomic profiles of adult patients with brain DMG.
Materials and Methods
Patients aged ≥ 18 years diagnosed with brain DMG at Seoul National University Hospital were included. The clinicopathological parameters, treatment outcomes, survival, and genomic profiles using 82-gene targeted next-generation sequencing (NGS) were analyzed. The 6-month progression-free survival (PFS6) after radiotherapy and overall survival (OS) were evaluated.
Results
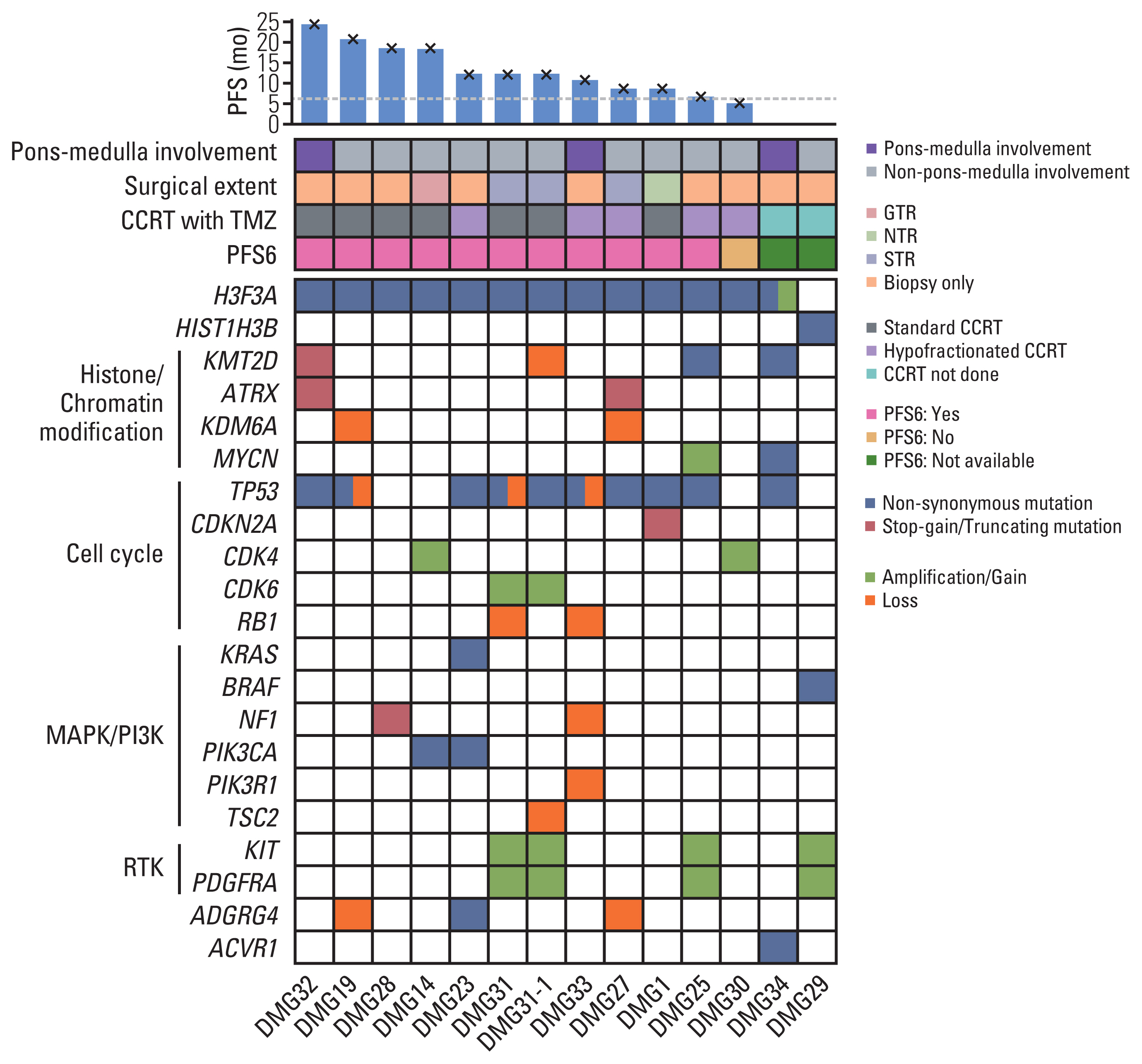
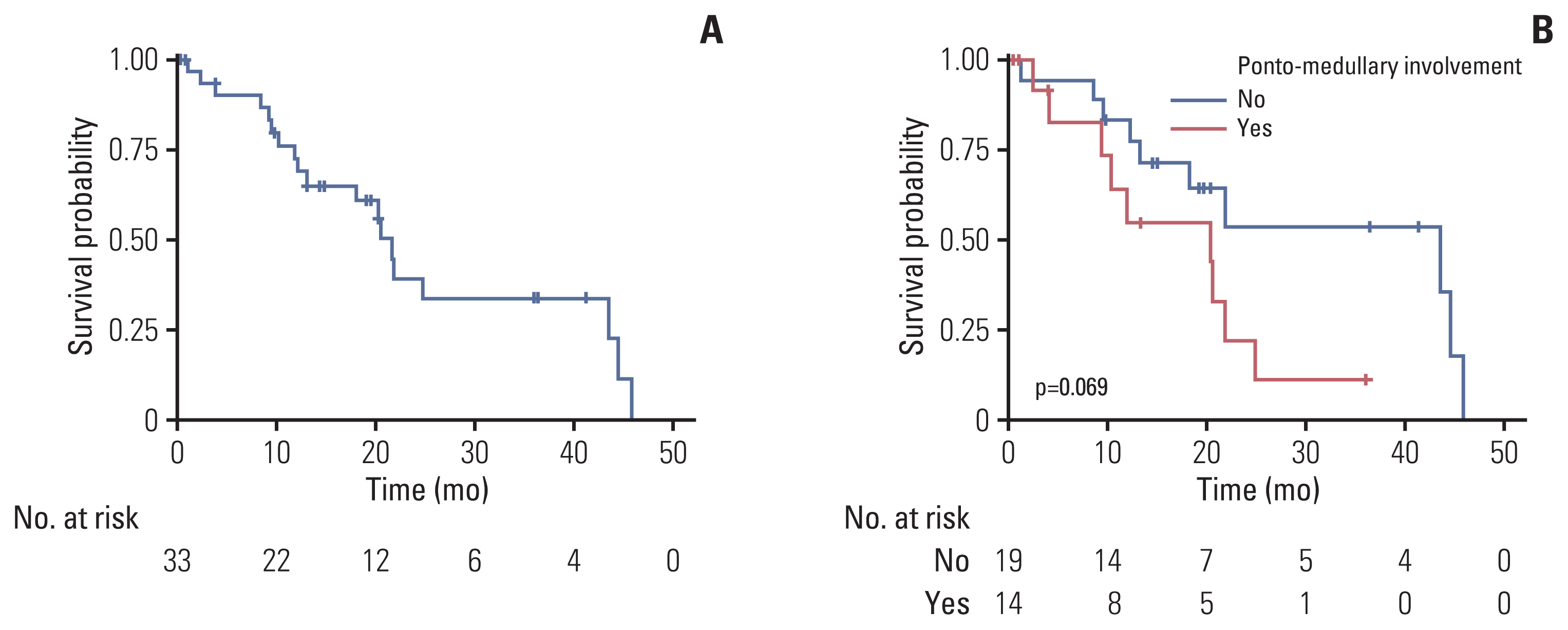
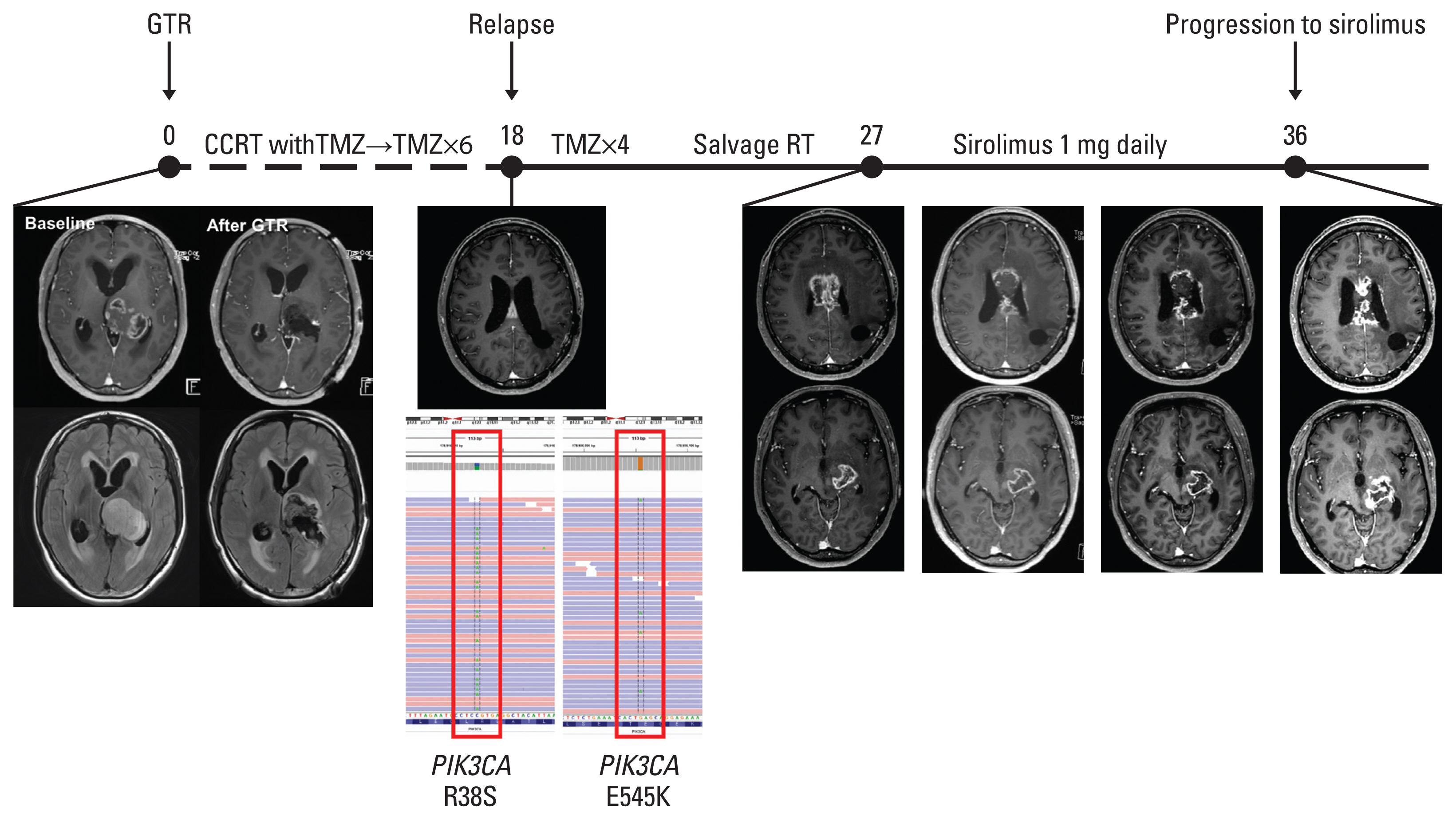
Thirty-three patients with H3-mutant brain DMG were identified. The median OS from diagnosis was 21.8 months (95% confidence interval [CI], 13.2 to not available [NA]) and involvement of the ponto-medullary area tended to have poor OS (median OS, 20.4 months [95% CI, 9.3 to NA] vs. 43.6 months [95% CI, 18.2 to NA]; p=0.07). Twenty-four patients (72.7%) received radiotherapy with or without temozolomide. The PFS6 rate was 83.3% (n=20). Patients without progression at 6 months showed significantly prolonged OS compared with those with progression at 6 months (median OS, 24.9 months [95% CI, 20.4 to NA] vs. 10.8 months [95% CI, 4.0 to NA]; p=0.02, respectively). Targeted NGS was performed in 13 patients with DMG, among whom nine (69.2%) harbored concurrent TP53 mutation. Two patients (DMG14 and DMG23) with PIK3CAR38S+E545K and KRASG12A mutations received matched therapies. Patient DMG14 received sirolimus with a PFS of 8.4 months.
Conclusion
PFS6 after radiotherapy was associated with prolonged survival in adult patients with DMG. Genome-based matched therapy may be an encouraging approach for progressive adult patients with DMG.
Figure
Cited by 1 articles
-
Choosing Wisely between Radiotherapy Dose-Fractionation Schedules: The Molecular Graded Prognostic Assessment for Elderly Glioblastoma Patients
Hye In Lee, Jina Kim, In Ah Kim, Joo Ho Lee, Jaeho Cho, Rifaquat Rahman, Geoffrey Fell, Chan Woo Wee, Hong In Yoon
Cancer Res Treat. 2025;57(2):378-386. doi: 10.4143/crt.2024.680.
Reference
-
References
1. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–20.
Article2. Gielen GH, Gessi M, Hammes J, Kramm CM, Waha A, Pietsch T. H3F3A K27M mutation in pediatric CNS tumors: a marker for diffuse high-grade astrocytomas. Am J Clin Pathol. 2013; 139:345–9.3. Karremann M, Gielen GH, Hoffmann M, Wiese M, Colditz N, Warmuth-Metz M, et al. Diffuse high-grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro Oncol. 2018; 20:123–31.
Article4. Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017; 32:520–37.
Article5. Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–31.6. Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014; 46:451–6.7. Aihara K, Mukasa A, Gotoh K, Saito K, Nagae G, Tsuji S, et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol. 2014; 16:140–6.
Article8. Meyronet D, Esteban-Mader M, Bonnet C, Joly MO, Uro-Coste E, Amiel-Benouaich A, et al. Characteristics of H3 K27M-mutant gliomas in adults. Neuro Oncol. 2017; 19:1127–34.
Article9. Rodon J, Soria JC, Berger R, Miller WH, Rubin E, Kugel A, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019; 25:751–8.
Article10. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005; 352:987–96.
Article11. Sulman EP, Ismaila N, Armstrong TS, Tsien C, Batchelor TT, Cloughesy T, et al. Radiation therapy for glioblastoma: American Society of Clinical Oncology clinical practice guideline endorsement of the American Society for Radiation Oncology guideline. J Clin Oncol. 2017; 35:361–9.
Article12. Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010; 28:1963–72.
Article13. Ballman KV, Buckner JC, Brown PD, Giannini C, Flynn PJ, LaPlant BR, et al. The relationship between six-month progression-free survival and 12-month overall survival end points for phase II trials in patients with glioblastoma multiforme. Neuro Oncol. 2007; 9:29–38.
Article14. Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012; 28:1838–44.
Article15. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43(11–10):1–33.16. Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–201.
Article17. Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012; 28:i333–9.
Article18. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016; 32:1220–2.
Article19. Oesper L, Satas G, Raphael BJ. Quantifying tumor heterogeneity in whole-genome and whole-exome sequencing data. Bioinformatics. 2014; 30:3532–40.
Article20. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016; 12:e1004873.
Article21. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92.22. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–45.
Article23. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019; 47:D941–7.
Article24. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–11.
Article25. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014; 42:D980–5.
Article26. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–91.27. Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016; 7:11185.
Article28. Schreck KC, Ranjan S, Skorupan N, Bettegowda C, Eberhart CG, Ames HM, et al. Incidence and clinicopathologic features of H3 K27M mutations in adults with radiographically-determined midline gliomas. J Neurooncol. 2019; 143:87–93.
Article29. Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019; 35:140–55.
Article30. Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013; 340:857–61.
Article31. Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013; 27:985–90.
Article32. Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012; 22:425–37.33. Jung KS, Lee J, Park SH, Park JO, Park YS, Lim HY, et al. Pilot study of sirolimus in patients with PIK3CA mutant/amplified refractory solid cancer. Mol Clin Oncol. 2017; 7:27–31.
Article34. Lin GL, Wilson KM, Ceribelli M, Stanton BZ, Woo PJ, Kreimer S, et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci Transl Med. 2019; 11:eaaw0064.
Article35. Yamaguchi H, Hung MC. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014; 46:209–22.
Article36. Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017; 23:483–92.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Features and Prognosis of Diffuse Midline Glioma: A Series of 24 Cases
- Diffuse Midline Gliomas Harboring the H3 K27M-Mutation in the Bilateral Thalamus and Midbrain: A Case Report and a Review of the Literature
- Epigenetic and Metabolic Changes in Diffuse Intrinsic Pontine Glioma
- The spectrum of microvascular patterns in adult diffuse glioma and their correlation with tumor grade
- Classification and Diagnosis of Adult Glioma: A Scoping Review