J Pathol Transl Med.
2021 Mar;55(2):102-111. 10.4132/jptm.2020.10.22.
The prognostic significance of p16 expression pattern in diffuse gliomas
- Affiliations
-
- 1Department of Pathology, Seoul National University Hospital, Seoul, Korea
- 2Department of Neurosurgery, Seoul National University Hospital, Seoul, Korea
- 3Neuroscience Research Institute, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2513895
- DOI: http://doi.org/10.4132/jptm.2020.10.22
Abstract
- Background
CDKN2A is a tumor suppressor gene that encodes the cell cycle inhibitor protein p16. Homozygous deletion of the CDKN2A gene has been associated with shortened survival in isocitrate dehydrogenase (IDH)–mutant gliomas. This study aimed to analyze the prognostic value of p16 and to evaluate whether p16 immunohistochemical staining could be used as a prognostic marker to replace CDKN2A genotyping in diffuse gliomas.
Methods
p16 immunohistochemistry was performed on tissue microarrays of 326 diffuse gliomas with diagnoses that reflected IDH-mutations and 1p/19q codeletion status. The results were divided into three groups (negative, focal expression, overexpression) according to the presence and degree of p16 expression. Survival analysis was performed to assess the prognostic value of p16 expression.
Results
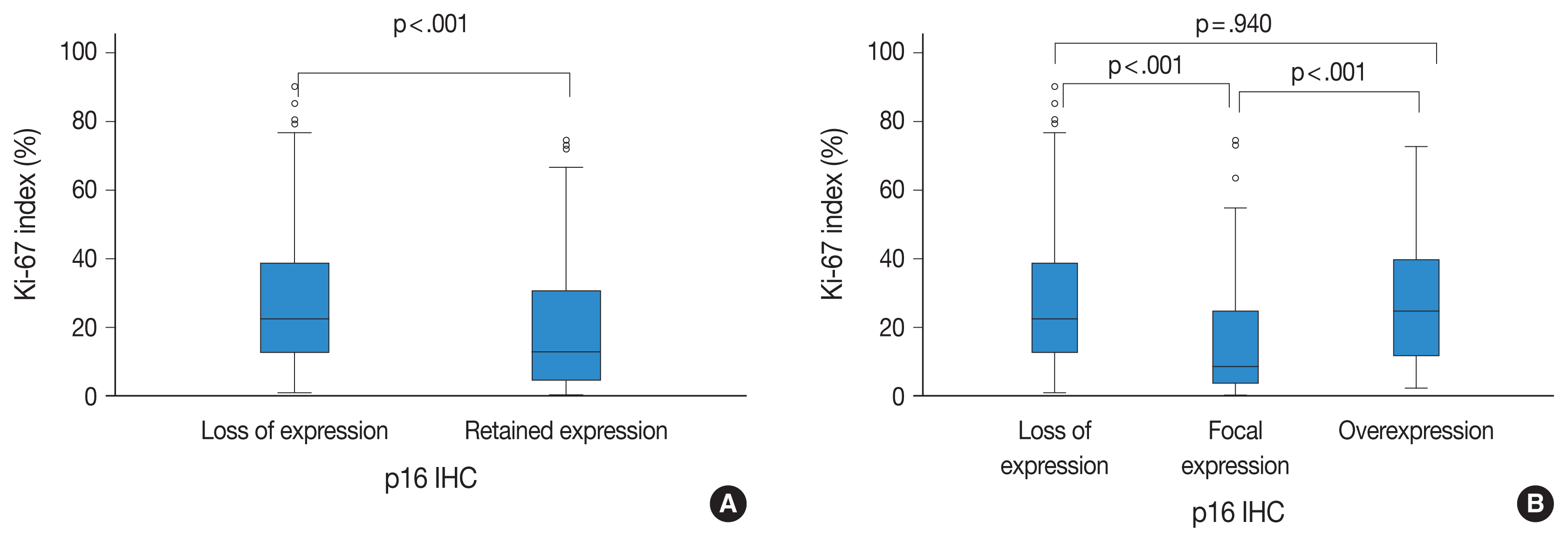
A loss of p16 expression predicted a significantly worse outcome in all glioma patients (n=326, p<.001), in the IDH-mutant glioma patients (n=103, p=.010), and in the IDH-mutant astrocytoma patients (n=73, p=.032). However, loss of p16 expression did not predict the outcome in the IDH-wildtype glioma patients (n=223, p=.121) or in the oligodendroglial tumor patients with the IDH-mutation and 1p/19q codeletion (n=30, p=.457). Multivariate analysis showed the association was still significant in the IDH-mutant glioma patients (p=.008; hazard ratio [HR], 2.637; 95% confidence interval [CI], 1.295 to 5.372) and in the IDH-mutant astrocytoma patients (p=.001; HR, 3.586; 95% CI, 1.649 to 7.801). Interestingly, patients who presented with tumors with p16 overexpression also had shorter survival times than did patients with tumors with p16 focal expression in the whole glioma (p< .001) and in IDH-mutant glioma groups. (p=.046).
Conclusions
This study suggests that detection of p16 expression by immunohistochemistry can be used as a useful surrogate test to predict prognosis, especially in IDH-mutant astrocytoma patients.
Keyword
Figure
Reference
-
References
1. Foulkes WD, Flanders TY, Pollock PM, Hayward NK. The CDKN2A (p16) gene and human cancer. Mol Med. 1997; 3:5–20.2. Serrano M. The tumor suppressor protein p16INK4a. Exp Cell Res. 1997; 237:7–13.
Article3. Raschke S, Balz V, Efferth T, Schulz WA, Florl AR. Homozygous deletions of CDKN2A caused by alternative mechanisms in various human cancer cell lines. Genes Chromosomes Cancer. 2005; 42:58–67.4. Appay R, Dehais C, Maurage CA, et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019; 21:1519–28.5. Reis GF, Pekmezci M, Hansen HM, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II–III) astrocytomas. J Neuropathol Exp Neurol. 2015; 74:442–52.6. Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018; 136:153–66.7. Yang RR, Shi ZF, Zhang ZY, et al. IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2020; 30:541–53.8. Aoki K, Nakamura H, Suzuki H, et al. Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 2018; 20:66–77.
Article9. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020; 139:603–8.10. Louis DN, Wesseling P, Aldape K, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020; 30:844–56.
Article11. Frazao L, do Carmo Martins M, Nunes VM, et al. BRAF V600E mutation and 9p21: CDKN2A/B and MTAP co-deletions: markers in the clinical stratification of pediatric gliomas. BMC Cancer. 2018; 18:1259.
Article12. Miettinen H, Kononen J, Sallinen P, et al. CDKN2/p16 predicts survival in oligodendrogliomas: comparison with astrocytomas. J Neurooncol. 1999; 41:205–11.13. Bortolotto S, Chiado-Piat L, Cavalla P, et al. CDKN2A/p16 inactivation in the prognosis of oligodendrogliomas. Int J Cancer. 2000; 88:554–7.
Article14. Kirla R, Salminen E, Huhtala S, et al. Prognostic value of the expression of tumor suppressor genes p53, p21, p16 and prb, and Ki-67 labelling in high grade astrocytomas treated with radiotherapy. J Neurooncol. 2000; 46:71–80.15. Jeon YK, Park K, Park CK, Paek SH, Jung HW, Park SH. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization. Neuropathology. 2007; 27:10–20.
Article16. Puduvalli VK, Kyritsis AP, Hess KR, et al. Patterns of expression of Rb and p16 in astrocytic gliomas, and correlation with survival. Int J Oncol. 2000; 17:963–9.
Article17. Arifin MT, Hama S, Kajiwara Y, et al. Cytoplasmic, but not nuclear, p16 expression may signal poor prognosis in high-grade astrocytomas. J Neurooncol. 2006; 77:273–7.
Article18. Milde-Langosch K, Bamberger AM, Rieck G, Kelp B, Loning T. Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat. 2001; 67:61–70.
Article19. O’Neill CJ, McCluggage WG. p16 expression in the female genital tract and its value in diagnosis. Adv Anat Pathol. 2006; 13:8–15.20. Lam AK, Ong K, Giv MJ, Ho YH. p16 expression in colorectal adenocarcinoma: marker of aggressiveness and morphological types. Pathology. 2008; 40:580–5.
Article21. Romagosa C, Simonetti S, Lopez-Vicente L, et al. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011; 30:2087–97.
Article22. Chaurasia A, Park SH, Seo JW, Park CK. Immunohistochemical analysis of ATRX, IDH1 and p53 in glioblastoma and their correlations with patient survival. J Korean Med Sci. 2016; 31:1208–14.
Article23. Koh J, Cho H, Kim H, et al. IDH2 mutation in gliomas including novel mutation. Neuropathology. 2015; 35:236–44.24. Kim SI, Lee Y, Won JK, Park CK, Choi SH, Park SH. Reclassification of mixed oligoastrocytic tumors using a genetically integrated diagnostic approach. J Pathol Transl Med. 2018; 52:28–36.
Article25. Chung CT, da Santos GC, Hwang DM, et al. FISH assay development for the detection of p16/CDKN2A deletion in malignant pleural mesothelioma. J Clin Pathol. 2010; 63:630–4.
Article26. Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011; 476:163–9.
Article27. Lotan TL, Wei W, Ludkovski O, et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol. 2016; 29:904–14.
Article28. Lu VM, O’Connor KP, Shah AH, et al. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol. 2020; 148:221–9.29. Burns KL, Ueki K, Jhung SL, Koh J, Louis DN. Molecular genetic correlates of p16, cdk4, and pRb immunohistochemistry in glioblastomas. J Neuropathol Exp Neurol. 1998; 57:122–30.
Article30. Purkait S, Jha P, Sharma MC, et al. CDKN2A deletion in pediatric versus adult glioblastomas and predictive value of p16 immunohistochemistry. Neuropathology. 2013; 33:405–12.31. Purkait S, Sharma V, Jha P, et al. EZH2 expression in gliomas: correlation with CDKN2A gene deletion/ p16 loss and MIB-1 proliferation index. Neuropathology. 2015; 35:421–31.32. Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol. 2002; 12:67–86.
Article33. Chiosea S, Krasinskas A, Cagle PT, Mitchell KA, Zander DS, Dacic S. Diagnostic importance of 9p21 homozygous deletion in malignant mesotheliomas. Mod Pathol. 2008; 21:742–7.
Article34. Piva R, Cavalla P, Bortolotto S, et al. CDKN2/p16 inactivation and p16 immunohistochemistry in astrocytic gliomas. Int J Oncol. 1998; 12:55–8.
Article35. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010; 17:510–22.
Article36. Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hyper-methylation of the RB1 gene in glioblastomas. Lab Invest. 2001; 81:77–82.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Expression of p16 in Diffuse Large B-cell Lymphoma and Its Prognostic Implications
- Expression of p16, Rb and FHIT Proteins in Urothelial Carcinoma of the Urinary Bladder
- Protein Phosphatase Magnesium-Dependent 1δ (PPM1D) Expression as a Prognostic Marker in Adult Supratentorial Diffuse Astrocytic and Oligodendroglial Tumors
- Expression of Trans forming Growth Factor-a and Proliferating Cell Nuclear Antigen in Human Gliomas
- The prognostic significance of p16, Ki-67, p63, and CK17 expression determined by immunohistochemical staining in cervical intraepithelial neoplasia 1