Ann Lab Med.
2021 Jul;41(4):401-408. 10.3343/alm.2021.41.4.401.
Clinical Application of Sequential Epigenetic Analysis for Diagnosis of Silver–Russell Syndrome
- Affiliations
-
- 1Pediatric Clinical Neuroscience Center, Department of Pediatrics, Seoul National University Children’s Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Division of Pediatric Orthopedics, Department of Orthopaedic Surgery, Seoul National University Children’s Hospital, Seoul National University College of Medicine, Seoul, Korea
- 3Division of Endocrinology, Department Pediatrics, Seoul National University Children’s Hospital, Seoul National University College of Medicine, Seoul, Korea
- 4Division of Clinical Genetics, Department of Pediatrics, Seoul National University Children’s Hospital, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2512720
- DOI: http://doi.org/10.3343/alm.2021.41.4.401
Abstract
- Background
Silver-Russell syndrome (SRS) is a pre- or post-natal growth retardation disorder caused by (epi)genetic alterations. We evaluated the molecular basis and clinical value of sequential epigenetic analysis in pediatric patients with SRS.
Methods
Twenty-eight patients who met ≥ 3 Netchine-Harbison clinical scoring system (NH-CSS) criteria for SRS were enrolled;26 (92.9%) were born small for gestational age, and 25 (89.3%) showed postnatal growth failure. Relative macrocephaly, body asymmetry, and feeding difficulty were noted in 18 (64.3%), 13 (46.4%), and 9 (32.1%) patients, respectively. Methylation-specific multiplex ligation-dependent probe amplification (MSMLPA) on chromosome 11p15 was performed as the first diagnostic step. Subsequently, bisulfite pyrosequencing (BP) for imprinting center 1 and 2 (IC1 and IC2) at chromosome 11p15, MEST on chromosome 7q32.2, and MEG3 on chromosome 14q32.2 was performed.
Results
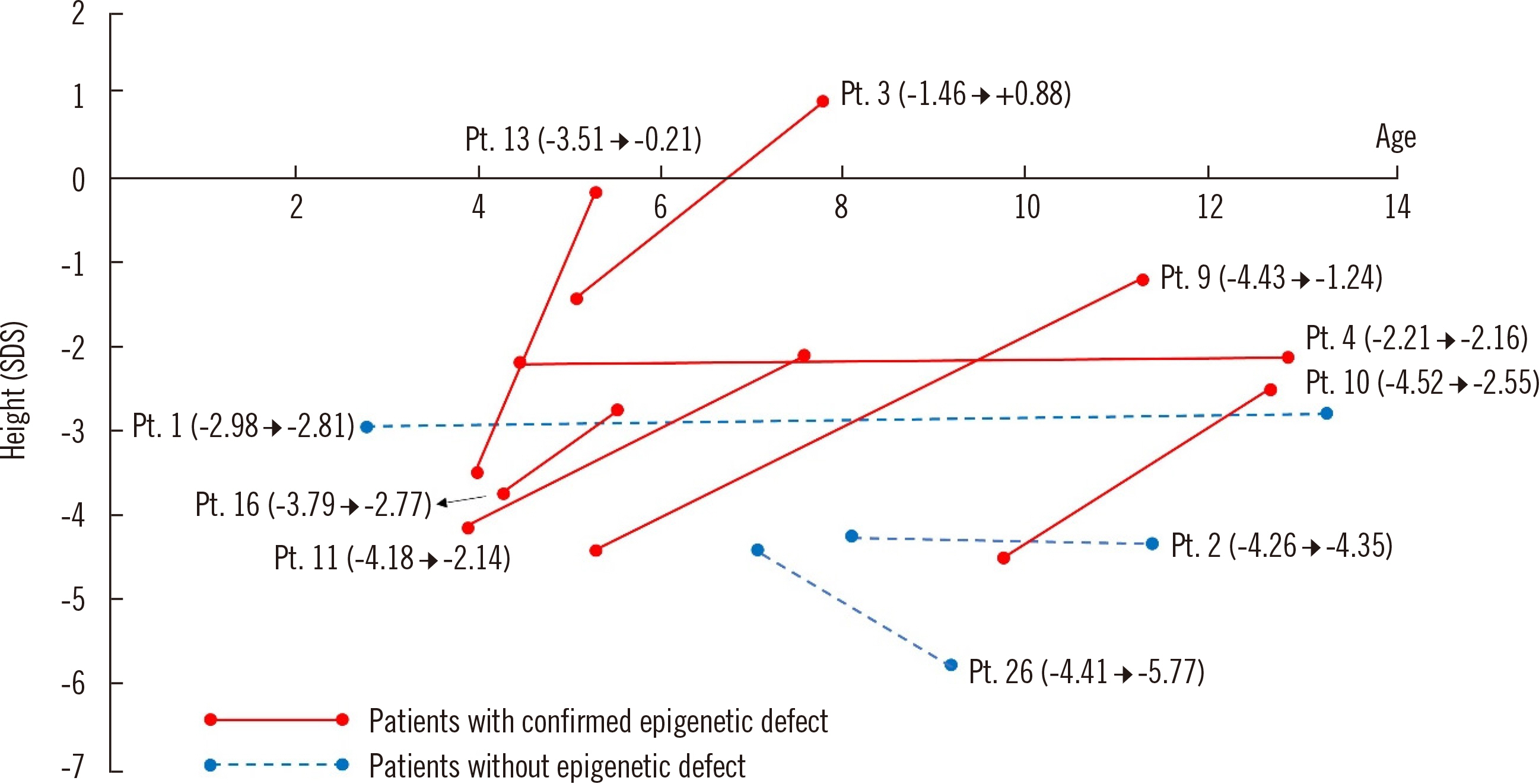
. Seventeen (60.7%) patients exhibited methylation defects, including loss of IC1 methylation (N = 14; 11 detected by MS-MLPA and three detected by BP) and maternal uniparental disomy 7 (N = 3). The diagnostic yield was comparable between patients who met three or four of the NH-CSS criteria (53.8% vs 50.0%). Patients with methylation defects responded better to growth hormone treatment.
Conclusions
NH-CSS is a powerful tool for SRS screening. However, in practice, genetic analysis should be considered even in patients with a low NH-CSS score. BP analysis detected additional methylation defects that were missed by MS-MLPA and might be considered as a first-line diagnostic tool for SRS.
Keyword
Figure
Reference
-
1. Silver HK, Kiyasu W, George J, Deamer WC. 1953; Syndrome of congenital hemihypertrophy, shortness of stature, and elevated urinary gonadotropins. Pediatrics. 12:368–76.2. Russell A. 1954; A syndrome of intra-uterine dwarfism recognizable at birth with cranio-facial dysostosis, disproportionately short arms, and other anomalies (5 examples). Proc R Soc Med. 47:1040–4.3. Wollmann HA, Kirchner T, Enders H, Preece MA, Ranke MB. 1995; Growth and symptoms in Silver-Russell syndrome: review on the basis of 386 patients. Eur J Pediatr. 154:958–68. DOI: 10.1007/BF01958638. PMID: 8801103.
Article4. Eggermann T. 2010; Russell-Silver syndrome. Am J Med Genet C Semin Med Genet. 154C:355–64. DOI: 10.1002/ajmg.c.30274. PMID: 20803658.
Article5. Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, et al. 2005; Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 37:1003–7. DOI: 10.1038/ng1629. PMID: 16086014.
Article6. Eggermann T, Schönherr N, Meyer E, Obermann C, Mavany M, Eggermann K, et al. 2006; Epigenetic mutations in 11p15 in Silver-Russell syndrome are restricted to the telomeric imprinting domain. J Med Genet. 43:615–6. DOI: 10.1136/jmg.2005.038687. PMID: 16236811. PMCID: PMC2564559.
Article7. Netchine I, Rossignol S, Dufourg MN, Azzi S, Rousseau A, Perin L, et al. 2007; 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab. 92:3148–54. DOI: 10.1210/jc.2007-0354. PMID: 17504900.
Article8. Kotzot D, Schmitt S, Bernasconi F, Robinson WP, Lurie IW, Ilyina H, et al. 1995; Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum Mol Genet. 4:583–7. DOI: 10.1093/hmg/4.4.583. PMID: 7633407.
Article9. Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE. 2008; The genetic aetiology of Silver-Russell syndrome. J Med Genet. 45:193–9. DOI: 10.1136/jmg.2007.053017. PMID: 18156438.
Article10. Cytrynbaum C, Chong K, Hannig V, Choufani S, Shuman C, Steele L, et al. 2016; Genomic imbalance in the centromeric 11p15 imprinting center in three families: Further evidence of a role for IC2 as a cause of Russell-Silver syndrome. Am J Med Genet A. 170:2731–9. DOI: 10.1002/ajmg.a.37819. PMID: 27374371.
Article11. Inoue T, Nakamura A, Fuke T, Yamazawa K, Sano S, Matsubara K, et al. 2017; Genetic heterogeneity of patients with suspected Silver-Russell syndrome: genome-wide copy number analysis in 82 patients without imprinting defects. Clin Epigenetics. 9:52. DOI: 10.1186/s13148-017-0350-6. PMID: 28515796. PMCID: PMC5433143.
Article12. Crippa M, Bonati MT, Calzari L, Picinelli C, Gervasini C, Sironi A, et al. 2019; Molecular etiology disclosed by array CGH in patients with Silver-Russell syndrome or similar phenotypes. Front Genet. 10:955. DOI: 10.3389/fgene.2019.00955. PMID: 31749829. PMCID: PMC6843062.
Article13. Brioude F, Oliver-Petit I, Blaise A, Praz F, Rossignol S, Le Jule M, et al. 2013; CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell-Silver syndrome. J Med Genet. 50:823–30. DOI: 10.1136/jmedgenet-2013-101691. PMID: 24065356.14. Begemann M, Zirn B, Santen G, Wirthgen E, Soellner L, Büttel HM, et al. 2015; Paternally inherited IGF2 mutation and growth restriction. N Engl J Med. 373:349–56. DOI: 10.1056/NEJMoa1415227. PMID: 26154720.15. Abi Habib W, Brioude F, Edouard T, Bennett JT, Lienhardt-Roussie A, Tixier F, et al. 2018; Genetic disruption of the oncogenic HMGA2-PLAG1-IGF2 pathway causes fetal growth restriction. Genet Med. 20:250–8. DOI: 10.1038/gim.2017.105. PMID: 28796236. PMCID: PMC5846811.
Article16. Luk HM, Ivan Lo FM, Sano S, Matsubara K, Nakamura A, Ogata T, et al. 2016; Silver-Russell syndrome in a patient with somatic mosaicism for upd(11)mat identified by buccal cell analysis. Am J Med Genet A. 170:1938–41. DOI: 10.1002/ajmg.a.37679. PMID: 27150791. PMCID: PMC5084779.
Article17. Eggermann T, Begemann M, Binder G, Spengler S. 2010; Silver-Russell syndrome: genetic basis and molecular genetic testing. Orphanet J Rare Dis. 5:19. DOI: 10.1186/1750-1172-5-19. PMID: 20573229. PMCID: PMC2907323.
Article18. Price SM, Stanhope R, Garrett C, Preece MA, Trembath RC. 1999; The spectrum of Silver-Russell syndrome: a clinical and molecular genetic study and new diagnostic criteria. J Med Genet. 36:837–42.19. Hitchins MP, Stanier P, Preece MA, Moore GE. 2001; Silver-Russell syndrome: a dissection of the genetic aetiology and candidate chromosomal regions. J Med Genet. 38:810–9. DOI: 10.1136/jmg.38.12.810. PMID: 11748303. PMCID: PMC1734774.
Article20. Azzi S, Salem J, Thibaud N, Chantot-Bastaraud S, Lieber E, Netchine I, et al. 2015; A prospective study validating a clinical scoring system and demonstrating phenotypical-genotypical correlations in Silver-Russell syndrome. J Med Genet. 52:446–53. DOI: 10.1136/jmedgenet-2014-102979. PMID: 25951829. PMCID: PMC4501172.
Article21. Luk HM, Yeung KS, Wong WL, Chung Brian HY, Tong Tony MF, Lo Ivan FM. 2016; Silver-Russell syndrome in Hong Kong. Hong Kong Med J. 22:526–33. DOI: 10.12809/hkmj154750. PMID: 27468965.
Article22. Wakeling EL, Brioude F, Lokulo-Sodipe O, O'Connell SM, Salem J, Bliek J, et al. 2017; Diagnosis and management of Silver-Russell syndrome: first international consensus statement. Nat Rev Endocrinol. 13:105–24. DOI: 10.1038/nrendo.2016.138. PMID: 27585961.
Article23. Kim JH, Yun S, Hwang SS, Shim JO, Chae HW, Lee YJ, et al. 2018; The 2017 Korean National Growth Charts for children and adolescents: development, improvement, and prospects. Korean J Pediatr. 61:135–49. DOI: 10.3345/kjp.2018.61.5.135. PMID: 29853938. PMCID: PMC5976563.
Article24. Lee BH, Kim GH, Oh TJ, Kim JH, Lee JJ, Choi SH, et al. 2013; Quantitative analysis of methylation status at 11p15 and 7q21 for the genetic diagnosis of Beckwith-Wiedemann syndrome and Silver-Russell syndrome. J Hum Genet. 58:604–10. DOI: 10.1038/jhg.2013.67. PMID: 23803580.
Article25. Fuke T, Mizuno S, Nagai T, Hasegawa T, Horikawa R, Miyoshi Y, et al. 2013; Molecular and clinical studies in 138 Japanese patients with Silver-Russell syndrome. PLoS One. 8:e60105. DOI: 10.1371/journal.pone.0060105. PMID: 23533668. PMCID: PMC3606247.
Article26. Russo S, Calzari L, Mussa A, Mainini E, Cassina M, Di Candia S, et al. 2016; A multi-method approach to the molecular diagnosis of overt and borderline 11p15.5 defects underlying Silver-Russell and Beckwith-Wiedemann syndromes. Clin Epigenetics. 8:23. DOI: 10.1186/s13148-016-0183-8. PMID: 26933465. PMCID: PMC4772365.
Article27. Wakeling EL, Amero SA, Alders M, Bliek J, Forsythe E, Kumar S, et al. 2010; Epigenotype-phenotype correlations in Silver-Russell syndrome. J Med Genet. 47:760–8. DOI: 10.1136/jmg.2010.079111. PMID: 20685669. PMCID: PMC2976034.
Article28. Jang W, Kim Y, Han E, Park J, Chae H, Kwon A, et al. 2019; Chromosomal microarray analysis as a first-tier clinical diagnostic test in patients with developmental delay/intellectual disability, autism spectrum disorders, and multiple congenital anomalies: A prospective multicenter study in Korea. Ann Lab Med. 39:299–310. DOI: 10.3343/alm.2019.39.3.299. PMID: 30623622. PMCID: PMC6340852.
Article29. Lee JS, Hwang H, Kim SY, Kim KJ, Choi JS, Woo MJ, et al. 2018; Chromosomal microarray with clinical diagnostic utility in children with developmental delay or intellectual disability. Ann Lab Med. 38:473–80. DOI: 10.3343/alm.2018.38.5.473. PMID: 29797819. PMCID: PMC5973923.
Article30. Chernausek SD, Breen TJ, Frank GR. 1996; Linear growth in response to growth hormone treatment in children with short stature associated with intrauterine growth retardation: the National Cooperative Growth study experience. J Pediatr. 128:S22–7. DOI: 10.1016/S0022-3476(96)70006-9.
Article31. Toumba M, Albanese A, Azcona C, Stanhope R. 2010; Effect of long-term growth hormone treatment on final height of children with Russell-Silver syndrome. Horm Res Paediatr. 74:212–7. DOI: 10.1159/000295924. PMID: 20424422.
Article32. Binder G, Liebl M, Woelfle J, Eggermann T, Blumenstock G, Schweizer R. 2013; Adult height and epigenotype in children with Silver-Russell syndrome treated with GH. Horm Res Paediatr. 80:193–200. DOI: 10.1159/000354658. PMID: 24051620.
Article33. Rakover Y, Dietsch S, Ambler GR, Chock C, Thomsett M, Cowell CT. 1996; Growth hormone therapy in Silver-Russell syndrome: 5 years experience of the Australian and New Zealand Growth database (OZGROW). Eur J Pediatr. 155:851–7. DOI: 10.1007/BF02282833. PMID: 8891553.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Russell-Silver Syndrome
- Growth Hormone Therapy in Russell-Silver Syndrome
- Molecular characterization in chromosome 11p15.5 related imprinting disorders Beckwith–Wiedemann and Silver–Russell syndromes
- Prosthetic management of a growing patient with Russell-Silver syndrome: a clinical report
- Bone Age Determination and Hand Radiographic Findings in Children With Russell-Silver Syndrome