Ann Lab Med.
2021 May;41(3):285-292. 10.3343/alm.2021.41.3.285.
Prevalence and Molecular Epidemiology of ExtendedSpectrum-β-Lactamase (ESBL)-Producing Escherichia coli From Multiple Sectors of the Swine Industry in Korea: A Korean Nationwide Monitoring Program for a One Health Approach to Combat Antimicrobial Resistance
- Affiliations
-
- 1Department of Laboratory Medicine, National Health Insurance Service Ilsan Hospital, Goyang, Korea
- 2Department of Laboratory Medicine, National Police Hospital, Seoul, Korea
- 3Research Institute of Bacterial Resistance, Korea
- 4Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea
- 5Seoul Clinical Laboratories Academy, Yongin, Korea
- KMID: 2512686
- DOI: http://doi.org/10.3343/alm.2021.41.3.285
Abstract
- Background
One health is a flexible concept with many facets, including the environment, community, and the nosocomial super-bacteria resistance network. We investigated the molecular prevalence of extended-spectrum-β-lactamase-producing Escherichia coli (ESBL-EC) in workers, livestock, and the farm environment in Korea.
Methods
ESBL-EC isolates were obtained from samples from 19 swine farms, 35 retail stores, seven slaughterhouses, and 45 related workers throughout Korea from August 2017 to July 2018, using ChromID ESBL (BioMérieux, Marcy l’Etoile, France) agar and enrichment broth. The presence of ESBL and mobilized colistin resistance (mcr) genes and antimicrobial resistance were determined. Clonality was evaluated with pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST).
Results
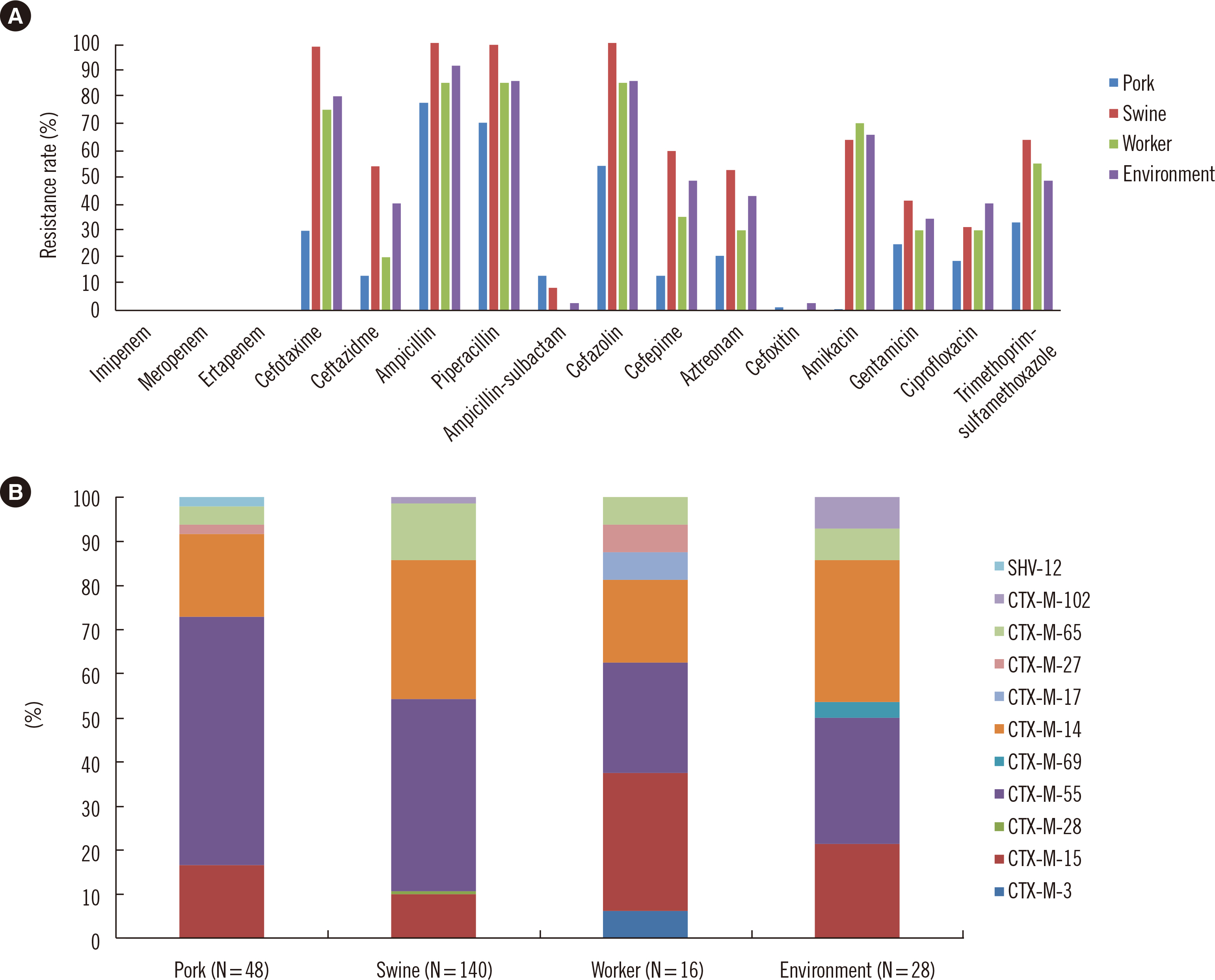
In total, 232 ESBL-EC isolates were obtained from 1,614 non-duplicated samples (14.4% positive rate). The ESBL-EC isolates showed regional and source-related differences. blaCTX-M-55 (N = 100), blaCTX-M-14 (N = 65), blaCTX-M-15 (N = 33), and blaCTX-M-65 (N = 23) were common ESBL types. The ESBL-EC isolates showed high resistance rates for various antimicrobial classes; however, all isolates were susceptible to carbapenem. One swine-originating colistin-resistant isolate did not carry any known mcr gene. PFGE was successful for 197 of the 232 isolates, and most PFGE types were heterogeneous, except for some dominant PFGE types (O, R, T, U, and V). MLST of 88 isolates was performed for representative PFGE types; however, no dominant sequence type was observed.
Conclusions
The proportion of ESBL-EC in swine industry-related samples was significant, and the isolates harbored common clinical ESBL gene types. These molecular epidemiologic data could provide important evidence for antimicrobial-resistance control through a one health approach.
Keyword
Figure
Reference
-
1. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. 2018; Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from Kor-GLASS. Euro Surveill. 23:1800047. DOI: 10.2807/1560-7917.ES.2018.23.42.1800047. PMID: 30352640. PMCID: PMC6199864.
Article2. Powell N, Davidson I, Yelling P, Collinson A, Pollard A, Johnson L, et al. 2017; Developing a local antimicrobial resistance action plan: the Cornwall One Health Antimicrobial Resistance Group. J Antimicrob Chemother. 72:2661–5. DOI: 10.1093/jac/dkx164. PMID: 28595316. PMCID: PMC5890714.
Article3. Na SH, Moon DC, Choi MJ, Oh SJ, Jung DY, Sung EJ, et al. 2019; Antimicrobial resistance and molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolated from ducks in South Korea. Foodborne Pathog Dis. 16:799–806. DOI: 10.1089/fpd.2019.2644. PMID: 31305137.4. CLSI. 2017. Performance standards for antimicrobial susceptibility testing. CLSI M100. 27th ed. Clinical and Laboratory Standards Institute;Wayne, PA:5. Ryoo NH, Kim E-C, Hong SG, Park YJ, Lee K, Bae IK, et al. 2005; Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 56:698–702. DOI: 10.1093/jac/dki324. PMID: 16141280.6. Rojas LJ, Salim M, Cober E, Richter SS, Perez F, Salata RA, et al. 2017; Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis. 64:711–8. DOI: 10.1093/cid/ciw805. PMID: 27940944. PMCID: PMC5850634.7. Yoon EJ, Hong JS, Yang JW, Lee KJ, Lee H, Jeong SH. 2018; Detection of mcr-1 plasmids in Enterobacteriaceae isolates from human specimens: comparison with those in Escherichia coli isolates from livestock in Korea. Ann Lab Med. 38:555–62. DOI: 10.3343/alm.2018.38.6.555. PMID: 30027699. PMCID: PMC6056395.8. Park YS, Bae IK, Kim J, Jeong SH, Hwang SS, Seo YH, et al. 2014; Risk factors and molecular epidemiology of community-onset extended-spectrum β-lactamase-producing Escherichia coli bacteremia. Yonsei Med J. 55:467–75. DOI: 10.3349/ymj.2014.55.2.467. PMID: 24532519. PMCID: PMC3936615.9. EnteroBase. Escherichia coli MLST database. https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search. Updated on Oct 2020.10. Andersson DI, Hughes D. 2014; Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 12:465–78. DOI: 10.1038/nrmicro3270. PMID: 24861036.
Article11. Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. 2018; Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 23:795. DOI: 10.3390/molecules23040795. PMID: 29601469. PMCID: PMC6017557.
Article12. Rousham EK, Unicomb L, Islam MA. 2018; Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc Biol Sci. 285:20180332. DOI: 10.1098/rspb.2018.0332. PMID: 29643217. PMCID: PMC5904322.
Article13. Lim S, Lee J, Lee H, Nam H, Moon D, Jang G, et al. 2014; Trends in antimicrobial sales for livestock and fisheries in Korea during 2003-2012. Korean J Vet Res. 54:81–6. DOI: 10.14405/kjvr.2014.54.2.81.
Article14. Lazarus B, Paterson DL, Mollinger JL, Rogers BA. 2015; Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. 60:439–52. DOI: 10.1093/cid/ciu785. PMID: 25301206.15. Kim H, Kim YA, Park YS, Choi MH, Lee GI, Lee K. 2017; Risk factors and molecular features of sequence type (ST) 131 extended-spectrum β-lactamase-producing Escherichia coli in community-onset bacteremia. Sci Rep. 7:14640. DOI: 10.1038/s41598-017-14621-4. PMID: 29116143. PMCID: PMC5677100.
Article16. Kim YA, Kim JJ, Kim H, Lee K. 2017; Community-onset extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 at two Korean community hospitals: The spread of multidrug-resistant E. coli to the community via healthcare facilities. Int J Infect Dis. 54:39–42. DOI: 10.1016/j.ijid.2016.11.010. PMID: 27865830.17. Kim YA, Kim H, Choi MH, Seo YH, Lee H, Lee K. 2020; Whole-genome analysis of blaCTX-M-55-carrying Escherichia coli among pigs, farm environment, and farm workers. Ann Lab Med. 40:180–3. DOI: 10.3343/alm.2020.40.2.180. PMID: 31650737. PMCID: PMC6822000.18. Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS 2nd, Hansen G, et al. 2012; Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 56:2364–70. DOI: 10.1128/AAC.05824-11. PMID: 22354301. PMCID: PMC3346636.19. Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, et al. 2014; Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli Sequence Type 131. J Clin Microbiol. 52:1358–65. DOI: 10.1128/JCM.03502-13. PMID: 24501035. PMCID: PMC3993632.20. Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011; Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol. 153:99–108. DOI: 10.1016/j.vetmic.2011.05.007. PMID: 21658865.21. Zurfluh K, Hächler H, Nüesch-Inderbinen M, Stephan R. 2013; Characteristics of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol. 79:3021–6. DOI: 10.1128/AEM.00054-13. PMID: 23455339. PMCID: PMC3623138.
Article22. Kim YA, Qureshi ZA, Adams-Haduch JM, Park YS, Shutt KA, Doi Y. 2012; Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin Infect Dis. 55:224–31. DOI: 10.1093/cid/cis387. PMID: 22491340. PMCID: PMC3491773.23. Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, et al. 2017; Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2:16260. DOI: 10.1038/nmicrobiol.2016.260. PMID: 28165472.
Article24. Fischer J, San José M, Roschanski N, Schmoger S, Baumann B, Irrgang A, et al. 2017; Spread and persistence of VIM-1 carbapenemase-producing Enterobacteriaceae in three German swine farms in 2011 and 2012. Vet Microbiol. 200:118–23. DOI: 10.1016/j.vetmic.2016.04.026. PMID: 27234907.
Article25. Hong JS, Song W, Jeong SH. 2020; Molecular characteristics of NDM-5-producing Escherichia coli from a cat and a dog in South Korea. Microb Drug Resist. 26:1005–8. DOI: 10.1089/mdr.2019.0382. PMID: 32043911.26. Lee JY, Lim SK, Choi Y, Moon DC, Shin J, Ko KS. 2018; Whole sequences and characteristics of mcr-1-harboring plasmids of Escherichia coli strains isolated from livestock in South Korea. Microb Drug Resist. 24:489–92. DOI: 10.1089/mdr.2017.0369. PMID: 29485936.27. Belaynehe KM, Shin SW, Park KY, Jang JY, Won HG, Yoon IJ, et al. 2018; Emergence of mcr-1 and mcr-3 variants coding for plasmid-mediated colistin resistance in Escherichia coli isolates from food- producing animals in South Korea. Int J Infect Dis. 72:22–4. DOI: 10.1016/j.ijid.2018.05.011. PMID: 29803875.28. Lim SK, Kang HY, Lee K, Moon DC, Lee HS, Jung SC. 2016; First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrob Agents Chemother. 60:6991–3. DOI: 10.1128/AAC.01472-16. PMID: 27572390. PMCID: PMC5075127.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antibiogram of Escherichia coli and Klebsiella spp. Detected by Vitek ESBL Test
- In Vitro Susceptibility of piperacillin/tazobactam Against extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae
- Molecular characteristics of ESBLproducing Escherichia coli isolated from chickens with colibacillosis
- Molecular Characteristics of Extended Spectrum beta-Lactamases in Escherichia coli and Klebsiella pneumoniae and the Prevalence of qnr in Extended Spectrum beta-Lactamase Isolates in a Tertiary Care Hospital in Korea
- Antimicrobial Susceptibility of Escherichia coli and Klebsiella pneumoniae Blood Isolates over 5 years:Influence of Extended-Spectrum Beta-Lactamase-Producing Organisms