Lab Med Online.
2020 Apr;10(2):169-174. 10.3343/lmo.2020.10.2.169.
Characteristics of Interstitial Deletion in Chromosome 4q Confirmed by Array Comparative Genomic Hybridization: A Case Report and Literature Review
- Affiliations
-
- 1Department of Pediatrics, Inje University Busan Paik Hospital, Busan, Korea
- 2Department of Radiology, Inje University Busan Paik Hospital, Busan, Korea
- 3Department of Laboratory Medicine, Inje University Busan Paik Hospital, Busan, Korea
- 4Department of Laboratory Medicine, Inje University Haeundae Paik Hospital, Busan, Korea
- KMID: 2512249
- DOI: http://doi.org/10.3343/lmo.2020.10.2.169
Abstract
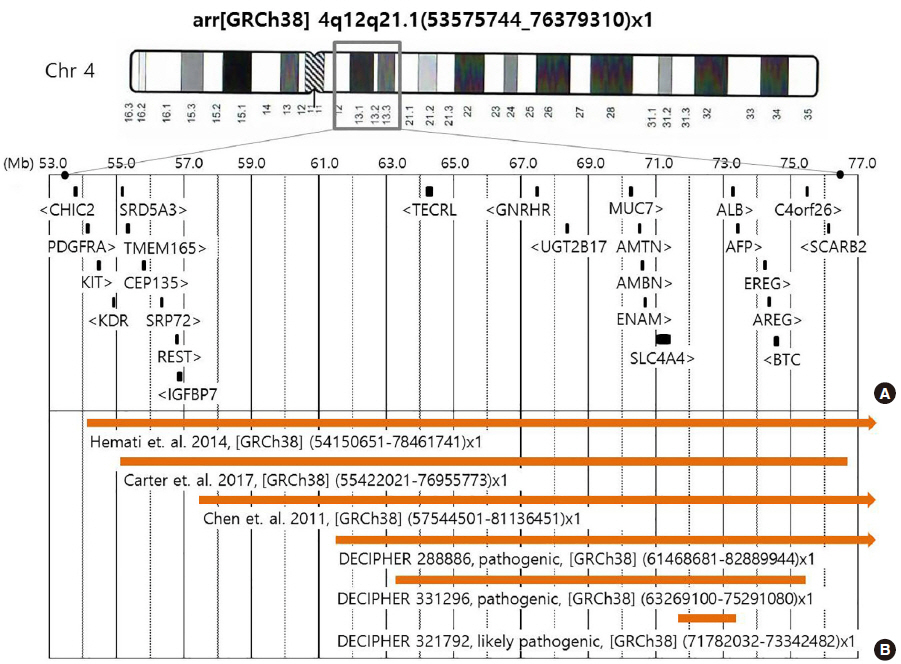
- Chromosome 4q deletion syndrome is a rare disease caused by partial deletion of the long arm of chromosome 4. Phenotypic severity and expressivity vary among patients with chromosome 4q deletions, depending on the size and region of the deletion of the affected chromosome. Although there have been many reports of proximal 4q deletion cases, very few have been confirmed by high-resolution array comparative genomic hybridization (aCGH). In the current study, we presented a new case of 4q proximal deletion, with detailed genetic and clinical characteristics, and compared these characteristics to those of six previous cases with available aCGH data. According to our review, several genes known to be associated with specific phenotypes of 4q12q21.1 deletion cannot sufficiently explain the variable phenotypes observed among the cases. These phenotypes include mental retardation, microcephaly, ocular anomalies, dental anomaly, and piebaldism. Consequently, we recommend further detailed investigations into the genes associated with 4q12q21.1 deletion to assist in identifying genotype-phenotype associations more clearly.
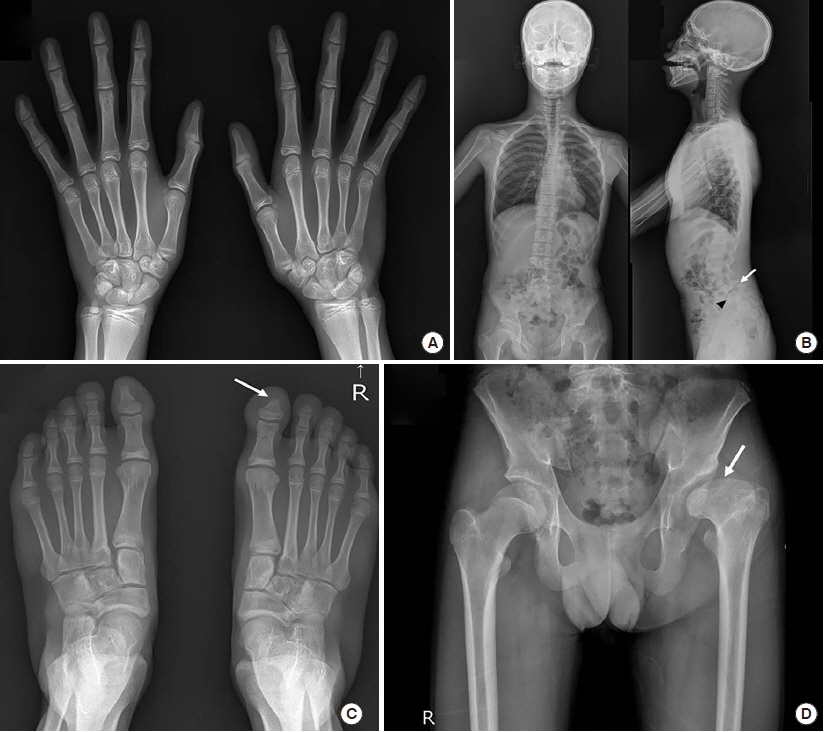
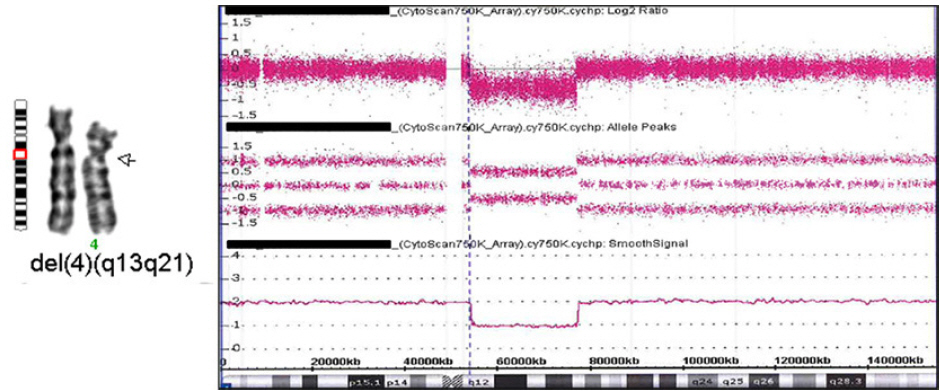
Figure
Reference
-
1. Strehle EM, Yu L, Rosenfeld JA, Donkervoort S, Zhou Y, Chen TJ, et al. 2012; Genotype-phenotype analysis of 4q deletion syndrome: Proposal of a critical region. Am J Med Genet A. 158A:2139–51. DOI: 10.1002/ajmg.a.35502. PMID: 22847869.
Article2. Komlósi K, Duga B, Hadzsiev K, Czakó M, Kosztolányi G, Fogarasi A, et al. 2015; Phenotypic variability in a Hungarian patient with the 4q21 microdeletion syndrome. Mol Cytogenet. 8:16. DOI: 10.1186/s13039-015-0118-7. PMID: 25774221. PMCID: PMC4359765.
Article3. Isamail S, Helmy NA, Mahmoud WM, El-Ruby MO. 2012; Phenotypic char-4 characterization of rare interstitial deletion of chromosome 4. J Pediatr Genet. 1:189–94. DOI: 10.3233/PGE-2012-029. PMID: 27625821. PMCID: PMC5020940.4. Strehle EM, Bantock HM. 2003; The phenotype of patients with 4q-syndrome. Genet Couns. 14:195–205. PMID: 12872814.5. Capalbo A, Sinibaldi L, Bernardini L, Spasari I, Mancuso B, Maggi E, et al. 2013; Interstitial 4q deletion associated with a mosaic complementary supernumerary marker chromosome in prenatal diagnosis. Prenat Diagn. 33:782–96. DOI: 10.1002/pd.4105. PMID: 23712311.
Article6. Kahrizi K1, Hu CH, Garshasbi M, Abedini SS, Ghadami S, Kariminejad R, et al. 2011; Next generation sequencing in a family with autosomal recessive Kahrizi syndrome (OMIM 612713) reveals a homozygous frameshift mutation in SRD5A3. Eur J Hum Genet. 19:115–7. DOI: 10.1038/ejhg.2010.132. PMID: 20700148. PMCID: PMC3039499.7. Igarashi T, Inatomi J, Sekine T, Cha SH, Kanai Y, Kunimi M, et al. 1999; Mu-tations in SLC4A4 cause permanent isolated proximal renal tubular acidosis with ocular abnormalities. Nat Genet. 23:264–6. DOI: 10.1038/15440. PMID: 10545938.8. Hemati P, du Souich C, Boerkoel CF. 2015; 4q12-4q21.21 deletion genotypephenotype correlation and the absence of piebaldism in presence of KIT haploinsufciency. Am J Med Genet A. 167A:231–7. DOI: 10.1002/ajmg.a.36821. PMID: 25355368.9. Carter J, Brittain H, Morrogh D, Lench N, Waters JJ. 2017; An interstitial 4q deletion with a mosaic complementary ring chromosome in a child with dysmorphism, linear skin pigmentation, and hepatomegaly. Case Rep Genet. 2017:4894515. DOI: 10.1155/2017/4894515. PMID: 28819573. PMCID: PMC5551472.
Article10. Chen CP, Lin SP, Su YN, Chern SR, Tsai FJ, Wu PC, et al. 2011; A 24.2-Mb deletion of 4q12 --> q21.21 characterized by array CGH in a 13½-year-old girl with short stature, mental retardation, developmental delay, hyperopia, exotropia, enamel defects, delayed tooth eruption and delayed puberty. Genet Couns. 22:255–61.11. Hussain MS, Baig SM, Neumann S, Nürnberg G, Farooq M, Ahmad I, et al. 2012; A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet. 90:871–8. DOI: 10.1016/j.ajhg.2012.03.016. PMID: 22521416. PMCID: PMC3376485.12. Farooq M, Fatima A, Mang Y, Hansen L, Kjaer KW, Baig SM, et al. 2016; A novel splice site mutation in CEP135 is associated with primary microcephaly in a Pakistani family. J Hum Genet. 61:271–3. DOI: 10.1038/jhg.2015.138. PMID: 26657937.13. Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, et al. 2005; ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 84:278–82. DOI: 10.1177/154405910508400314. PMID: 15723871.
Article14. Crawford PJ, Aldred M, Bloch-Zupan A. 2007; Amelogenesis imperfecta. Orphanet J Rare Dis. 2:17. DOI: 10.1186/1750-1172-2-17. PMID: 17408482. PMCID: PMC1853073.
Article15. Smith CE, Murillo G, Brookes SJ, Poulter JA, Silva S, Kirkham J, et al. 2016; Deletion of amelotin exons 3-6 is associated with amelogenesis imperfecta. Hum Mol Genet. 25:3578–87. DOI: 10.1093/hmg/ddw203. PMID: 27412008. PMCID: PMC5179951.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of an Interstitial Deletion in Chromosome 1p Confirmed by Array Comparative Genome Hybridization
- A de novo Proximal 6q Deletion Confirmed by Array Comparative Genomic Hybridization
- Terminal Deletion of the Chromosome 4q with Hemivertebra: Case Report
- A Case of 4q Deletion with Partial Agenesis of Corpus Callosum
- Current Status and Future Clinical Applications of Array.based Comparative Genomic Hybridization