Cancer Res Treat.
2020 Oct;52(4):1219-1228. 10.4143/crt.2019.688.
Evaluation of Circulating Tumor DNA in Patients with Ovarian Cancer Harboring Somatic PIK3CA or KRAS Mutations
- Affiliations
-
- 1Department of Gynecologic Oncology, Saitama Medical University International Medical Center, Hidaka, Japan
- 2Tsukuba Research Laboratories, Eisai Co., Ltd., Tsukuba, Japan
- KMID: 2507947
- DOI: http://doi.org/10.4143/crt.2019.688
Abstract
- Purpose
Circulating tumor DNA (ctDNA) is an attractive source for liquid biopsy to understand molecular phenotypes of a tumor non-invasively, which is also expected to be both a diagnostic and prognostic marker. PIK3CA and KRAS are among the most frequently mutated genes in epithelial ovarian cancer (EOC). In addition, their hotspot mutations have already been identified and are ready for a highly sensitive analysis. Our aim is to clarify the significance of PIK3CA and KRAS mutations in the plasma of EOC patients as tumor-informed ctDNA.
Methods
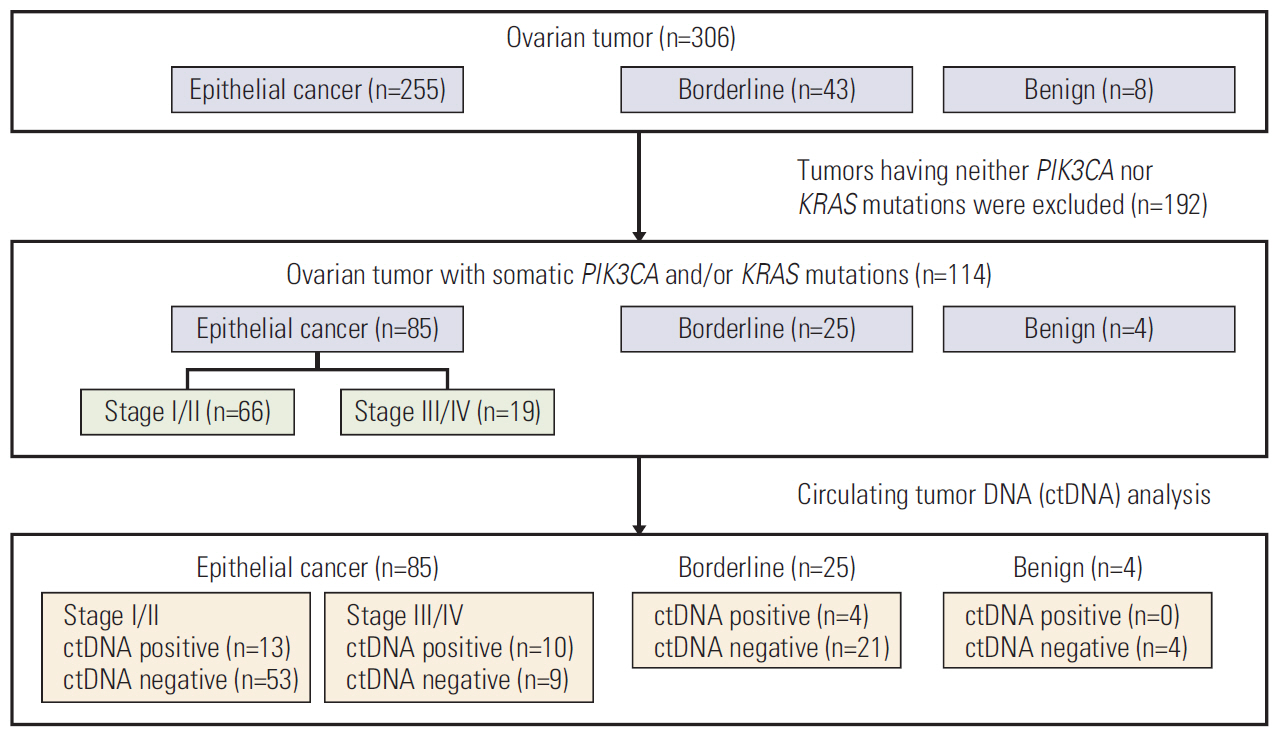
We screened 306 patients with ovarian tumors for somatic PIK3CA or KRAS mutations. A total of 85 EOC patients had somatic PIK3CA and/or KRAS mutations, and the corresponding mutations were subsequently analyzed using a droplet digital polymerase chain reaction in their plasma.
Results
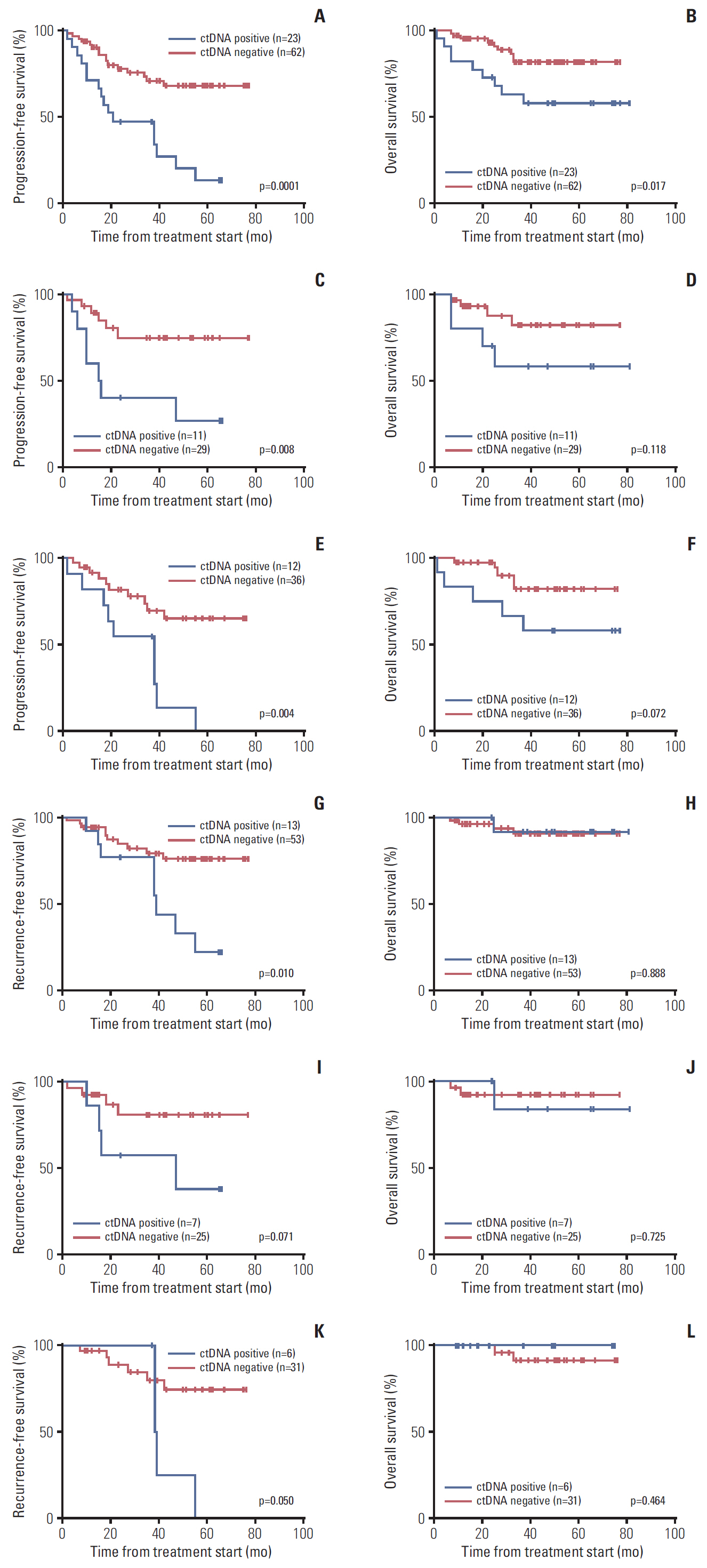
The detection rates for ctDNA were 27% in EOC patients. Advanced stage and positive peritoneal cytology were associated with higher frequency of ctDNA detection. Preoperative ctDNA detection was found to be an indicator of outcomes, and multivariate analysis revealed that ctDNA remained an independent risk factor for recurrence (p=0.010). Moreover, we assessed the mutation frequency in matched plasma before surgery and at recurrence from 17 patients, and found six patients had higher mutation rates in cell-free DNA at recurrence compared to that at primary diagnosis.
Conclusion
The presence of ctDNA at diagnosis was an indicator for recurrence, which suggests potential tumor spread even when tumors were localized at the time of diagnosis.
Keyword
Figure
Reference
-
References
1. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008; 14:985–90.
Article2. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010; 11:753–62.
Article3. Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015; 16:937–48.
Article4. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014; 4:650–61.
Article5. Mandel P, Metais P. Article in undetermined language. C R Seances Soc Biol Fil. 1948; 142:241–3.6. Offin M, Chabon JJ, Razavi P, Isbell JM, Rudin CM, Diehn M, et al. Capturing genomic evolution of lung cancers through liquid biopsy for circulating tumor DNA. J Oncol. 2017; 2017:4517834.
Article7. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013; 368:1199–209.
Article8. Cheuk IW, Shin VY, Kwong A. Detection of methylated circulating DNA as noninvasive biomarkers for breast cancer diagnosis. J Breast Cancer. 2017; 20:12–9.
Article9. Beije N, Helmijr JC, Weerts MJA, Beaufort CM, Wiggin M, Marziali A, et al. Somatic mutation detection using various targeted detection assays in paired samples of circulating tumor DNA, primary tumor and metastases from patients undergoing resection of colorectal liver metastases. Mol Oncol. 2016; 10:1575–84.
Article10. Przybyl J, Chabon JJ, Spans L, Ganjoo KN, Vennam S, Newman AM, et al. Combination approach for detecting different types of alterations in circulating tumor DNA in leiomyosarcoma. Clin Cancer Res. 2018; 24:2688–99.
Article11. Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer. 2016; 7:1907–14.
Article12. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.
Article13. Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009; 9:415–28.
Article14. Kim YM, Lee SW, Lee YJ, Lee HY, Lee JE, Choi EK. Prospective study of the efficacy and utility of TP53 mutations in circulating tumor DNA as a non-invasive biomarker of treatment response monitoring in patients with high-grade serous ovarian carcinoma. J Gynecol Oncol. 2019; 30:e32.
Article15. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017; 9:eaan2415.
Article16. Morikawa A, Hayashi T, Shimizu N, Kobayashi M, Taniue K, Takahashi A, et al. PIK3CA and KRAS mutations in cell free circulating DNA are useful markers for monitoring ovarian clear cell carcinoma. Oncotarget. 2018; 9:15266–74.
Article17. Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005; 193(3 Pt 1):662–7.
Article18. Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015; 10:e0145754.
Article19. Otsuka J, Okuda T, Sekizawa A, Amemiya S, Saito H, Okai T, et al. Detection of p53 mutations in the plasma DNA of patients with ovarian cancer. Int J Gynecol Cancer. 2004; 14:459–64.
Article20. Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016; 13:e1002198.
Article21. Zhuang R, Li S, Li Q, Guo X, Shen F, Sun H, et al. The prognostic value of KRAS mutation by cell-free DNA in cancer patients: a systematic review and meta-analysis. PLoS One. 2017; 12:e0182562.
Article22. Barbosa A, Peixoto A, Pinto P, Pinheiro M, Teixeira MR. Potential clinical applications of circulating cell-free DNA in ovarian cancer patients. Expert Rev Mol Med. 2018; 20:e6.
Article23. Bolivar AM, Luthra R, Mehrotra M, Chen W, Barkoh BA, Hu P, et al. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: identification of plasma-based, tumor-associated mutations in early stage patients. Mod Pathol. 2019; 32:405–14.
Article24. Scholer LV, Reinert T, Orntoft MW, Kassentoft CG, Arnadottir SS, Vang S, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017; 23:5437–45.
Article25. Iwama E, Sakai K, Azuma K, Harada T, Harada D, Nosaki K, et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol. 2017; 28:136–41.
Article26. Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat. 2015; 150:299–307.
Article27. Pietrasz D, Pecuchet N, Garlan F, Didelot A, Dubreuil O, Doat S, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res. 2017; 23:116–23.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Targeted Therapy for Pancreatic Cancer: Focus on KRAS
- Selective utilization of circulating tumor DNA testing enables disease monitoring in endometrial and ovarian carcinomas
- Analysis of Circulating Tumor DNA to Predict Neoadjuvant Therapy Effectiveness and Breast Cancer Recurrence
- Genomic mutation profiling using liquid biopsy in Korean patients with prostate cancer: Circulating tumor DNA mutation predicts the development of castration resistance
- Optimization to detect TP53 mutations in circulating cell-free tumor DNA from patients with serous epithelial ovarian cancer