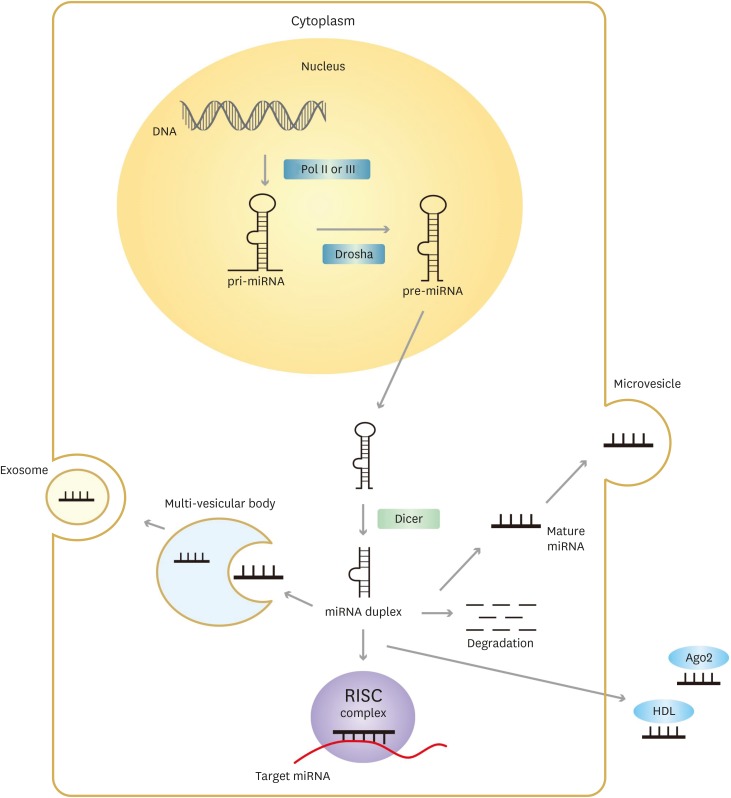
Infect Chemother.
2020 Mar;52(1):1-18. 10.3947/ic.2020.52.1.1.
An Update on Sepsis Biomarkers
- Affiliations
-
- 1Division of Infectious Disease, Department of Internal Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2507317
- DOI: http://doi.org/10.3947/ic.2020.52.1.1
Abstract
- Sepsis is a dysregulated systemic reaction to a common infection, that can cause lifethreatening organ dysfunction. Over the last decade, the mortality rate of patients with sepsis has decreased as long as patients are treated according to the recommendations of the Surviving Sepsis Campaign, but is still unacceptably high. Patients at risk of sepsis should therefore be identified prior to the onset of organ dysfunction and this requires a rapid diagnosis and a prompt initiation of treatment. Unfortunately, there is no gold standard for the diagnosis of sepsis and traditional standard culture methods are time-consuming. Recently, in order to overcome these limitations, biomarkers which could help in predicting the diagnosis and prognosis of sepsis, as well as being useful for monitoring the response to treatments, have been identified. In addition, recent advances have led to the development of newly identified classes of biomarkers such as microRNAs, long-non-coding RNAs, and the human microbiome. This review focuses on the latest information on biomarkers that can be used to predict the diagnosis and prognosis of sepsis.
Keyword
Figure
Reference
-
1. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016; 315:801–810. PMID: 26903338.
Article2. Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K. International Forum of Acute Care Trialists. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med. 2016; 193:259–272. PMID: 26414292.
Article3. Jeon K, Na SJ, Oh DK, Park S, Choi EY, Kim SC, Seong GM, Heo J, Chang Y, Kwack WG, Kang BJ, Choi WI, Kim KC, Park SY, Kwak SH, Shin YM, Lee HB, Park SH, Cho JH, Kim B, Lim CM. Korean Sepsis Alliance (KSA) study group. Characteristics, management and clinical outcomes of patients with sepsis: a multicenter cohort study in Korea. Acute Crit Care. 2019; 34:179–191. PMID: 31723927.
Article4. Ferreira FL, Bota DP, Bross A, Mélot C, Vincent JL. Serial evaluation of the SOFA score to predict outcome in critically ill patients. JAMA. 2001; 286:1754–1758. PMID: 11594901.
Article5. Arefian H, Heublein S, Scherag A, Brunkhorst FM, Younis MZ, Moerer O, Fischer D, Hartmann M. Hospital-related cost of sepsis: a systematic review. J Infect. 2017; 74:107–117. PMID: 27884733.
Article6. Rhee C, Dantes R, Epstein L, Murphy DJ, Seymour CW, Iwashyna TJ, Kadri SS, Angus DC, Danner RL, Fiore AE, Jernigan JA, Martin GS, Septimus E, Warren DK, Karcz A, Chan C, Menchaca JT, Wang R, Gruber S, Klompas M. CDC Prevention Epicenter Program. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA. 2017; 318:1241–1249. PMID: 28903154.
Article7. Esposito S, De Simone G, Boccia G, De Caro F, Pagliano P. Sepsis and septic shock: New definitions, new diagnostic and therapeutic approaches. J Glob Antimicrob Resist. 2017; 10:204–212. PMID: 28743646.
Article8. World Health Organization (WHO). WHO recommended surveillance standards. 2nd ed. 2004. 123–124. Accessed July 21, 2013. Available at: http://www.who.int/csr/resources/publications/surveillance/whocdscsrisr992.pdf.9. Jeon JH, Park DW. Controversies regarding the new definition of sepsis. Korean J Med. 2017; 92:342–348.
Article10. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001; 69:89–95. PMID: 11240971.11. Larsen FF, Petersen JA. Novel biomarkers for sepsis: A narrative review. Eur J Intern Med. 2017; 45:46–50. PMID: 28965741.
Article14. So-Ngern A, Leelasupasri S, Chulavatnatol S, Pummangura C, Bunupuradah P, Montakantikul P. Prognostic value of Serum Procalcitonin level for the diagnosis of bacterial infections in critically-ill patients. Infect Chemother. 2019; 51:263–273. PMID: 31583860.
Article15. Tian R, Wang X, Pan T, Li R, Wang J, Liu Z, Chen E, Mao E, Tan R, Chen Y, Liu J, Qu H. Plasma PTX3, MCP1 and Ang2 are early biomarkers to evaluate the severity of sepsis and septic shock. Scand J Immunol. 2019; 90:e12823. PMID: 31489646.
Article16. Bleharski JR, Kiessler V, Buonsanti C, Sieling PA, Stenger S, Colonna M, Modlin RL. A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J Immunol. 2003; 170:3812–3818. PMID: 12646648.
Article17. Gómez-Piña V, Soares-Schanoski A, Rodríguez-Rojas A, Del Fresno C, García F, Vallejo-Cremades MT, Fernández-Ruiz I, Arnalich F, Fuentes-Prior P, López-Collazo E. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J Immunol. 2007; 179:4065–4073. PMID: 17785845.
Article18. Khater WS, Salah-Eldeen NN, Khater MS, Saleh AN. Role of suPAR and lactic acid in diagnosing sepsis and predicting mortality in elderly patients. Eur J Microbiol Immunol (Bp). 2016; 6:178–185. PMID: 27766166.
Article19. Huang Q, Xiong H, Yan P, Shuai T, Liu J, Zhu L, Lu J, Yang K, Liu J. The diagnostic and prognostic value of supar in patients with sepsis: a systematic review and meta-analysis. Shock. 2020; 53:416–425. PMID: 31490358.
Article20. Yin WP, Li JB, Zheng XF, An L, Shao H, Li CS. Effect of neutrophil CD64 for diagnosing sepsis in emergency department. World J Emerg Med. 2020; 11:79–86. PMID: 32076472.
Article21. Ye Z, Zou H, Liu S, Mei C, Chang X, Hu Z, Yang H, Wu Y. Diagnostic performance of neutrophil CD64 index in patients with sepsis in the intensive care unit. J Int Med Res. 2019; 47:4304–4311. PMID: 31319721.
Article22. Yeh CF, Wu CC, Liu SH, Chen KF. Comparison of the accuracy of neutrophil CD64, procalcitonin, and C-reactive protein for sepsis identification: a systematic review and meta-analysis. Ann Intensive Care. 2019; 9:5. PMID: 30623257.
Article23. Sandquist M, Wong HR. Biomarkers of sepsis and their potential value in diagnosis, prognosis and treatment. Expert Rev Clin Immunol. 2014; 10:1349–1356. PMID: 25142036.
Article24. Yang HS, Hur M, Yi A, Kim H, Lee S, Kim SN. Prognostic value of presepsin in adult patients with sepsis: Systematic review and meta-analysis. PLoS One. 2018; 13:e0191486. PMID: 29364941.
Article25. Ulla M, Pizzolato E, Lucchiari M, Loiacono M, Soardo F, Forno D, Morello F, Lupia E, Moiraghi C, Mengozzi G, Battista S. Diagnostic and prognostic value of presepsin in the management of sepsis in the emergency department: a multicenter prospective study. Crit Care. 2013; 17:R168. PMID: 23899120.
Article26. Wu J, Hu L, Zhang G, Wu F, He T. Accuracy of presepsin in sepsis diagnosis: a systematic review and meta-analysis. PLoS One. 2015; 10:e0133057. PMID: 26192602.
Article27. Zhang J, Hu ZD, Song J, Shao J. Diagnostic value of presepsin for sepsis: a systematic review and meta-analysis. Medicine (Baltimore). 2015; 94:e2158. PMID: 26632748.28. Zhang X, Liu D, Liu YN, Wang R, Xie LX. The accuracy of presepsin (sCD14-ST) for the diagnosis of sepsis in adults: a meta-analysis. Crit Care. 2015; 19:323. PMID: 26357898.
Article29. Romualdo LG, Torrella PE, González MV, Sánchez RJ, Holgado AH, Freire AO, Acebes SR, Otón MD. Diagnostic accuracy of presepsin (soluble CD14 subtype) for prediction of bacteremia in patients with systemic inflammatory response syndrome in the Emergency Department. Clin Biochem. 2014; 47:505–508. PMID: 24560955.
Article30. Kweon OJ, Choi JH, Park SK, Park AJ. Usefulness of presepsin (sCD14 subtype) measurements as a new marker for the diagnosis and prediction of disease severity of sepsis in the Korean population. J Crit Care. 2014; 29:965–970. PMID: 25042676.
Article31. Lu B, Zhang Y, Li C, Liu C, Yao Y, Su M, Shou S. The utility of presepsin in diagnosis and risk stratification for the emergency patients with sepsis. Am J Emerg Med. 2018; 36:1341–1345. PMID: 29276032.
Article32. Godnic M, Stubljar D, Skvarc M, Jukic T. Diagnostic and prognostic value of sCD14-ST--presepsin for patients admitted to hospital intensive care unit (ICU). Wien Klin Wochenschr. 2015; 127:521–527. PMID: 25854904.
Article33. Yu H, Qi Z, Hang C, Fang Y, Shao R, Li C. Evaluating the value of dynamic procalcitonin and presepsin measurements for patients with severe sepsis. Am J Emerg Med. 2017; 35:835–841. PMID: 28153679.
Article34. Porte R, Davoudian S, Asgari F, Parente R, Mantovani A, Garlanda C, Bottazzi B. The long pentraxin PTX3 as a humoral innate immunity functional player and biomarker of infections and sepsis. Front Immunol. 2019; 10:794. PMID: 31031772.
Article35. Bottazzi B, Doni A, Garlanda C, Mantovani A. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annu Rev Immunol. 2010; 28:157–183. PMID: 19968561.
Article36. Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985; 13:818–829. PMID: 3928249.37. Le Gall JR, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA. 1993; 270:2957–2963. PMID: 8254858.
Article38. Hamed S, Behnes M, Pauly D, Lepiorz D, Barre M, Becher T, Lang S, Akin I, Borggrefe M, Bertsch T, Hoffmann U. Diagnostic value of Pentraxin-3 in patients with sepsis and septic shock in accordance with latest sepsis-3 definitions. BMC Infect Dis. 2017; 17:554. PMID: 28793880.
Article39. Lee YT, Gong M, Chau A, Wong WT, Bazoukis G, Wong SH, Lampropoulos K, Xia Y, Li G, Wong MCS, Liu T, Wu WKK, Tse G. International Heath Informatics Study (IHIS) Network. Pentraxin-3 as a marker of sepsis severity and predictor of mortality outcomes: a systematic review and meta-analysis. J Infect. 2018; 76:1–10. PMID: 29174966.
Article40. Hu C, Zhou Y, Liu C, Kang Y. Pentraxin-3, procalcitonin and lactate as prognostic markers in patients with sepsis and septic shock. Oncotarget. 2018; 9:5125–5136. PMID: 29435167.
Article41. Shabani F, Farasat A, Mahdavi M, Gheibi N. Calprotectin (S100A8/S100A9): a key protein between inflammation and cancer. Inflamm Res. 2018; 67:801–812. PMID: 30083975.
Article42. Larsson A, Tydén J, Johansson J, Lipcsey M, Bergquist M, Kultima K, Mandic-Havelka A. Calprotectin is superior to procalcitonin as a sepsis marker and predictor of 30-day mortality in intensive care patients. Scand J Clin Lab Invest. 2019; 1–6.
Article43. Huang L, Li J, Han Y, Zhao S, Zheng Y, Sui F, Xin X, Ma W, Jiang Y, Yao Y, Li W. Serum calprotectin expression as a diagnostic marker for sepsis in postoperative intensive care unit patients. J Interferon Cytokine Res. 2016; 36:607–616. PMID: 27610929.
Article44. Gao S, Yang Y, Fu Y, Guo W, Liu G. Diagnostic and prognostic value of myeloid-related protein complex 8/14 for sepsis. Am J Emerg Med. 2015; 33:1278–1282. PMID: 26206243.
Article45. Bartáková E, Štefan M, Stráníková A, Pospíšilová L, Arientová S, Beran O, Blahutová M, Máca J, Holub M. Calprotectin and calgranulin C serum levels in bacterial sepsis. Diagn Microbiol Infect Dis. 2019; 93:219–226. PMID: 30420210.
Article46. Jonsson N, Nilsen T, Gille-Johnson P, Bell M, Martling CR, Larsson A, Mårtensson J. Calprotectin as an early biomarker of bacterial infections in critically ill patients: an exploratory cohort assessment. Crit Care Resusc. 2017; 19:205–213. PMID: 28866970.47. Havelka A, Sejersen K, Venge P, Pauksens K, Larsson A. Calprotectin, a new biomarker for diagnosis of acute respiratory infections. Sci Rep. 2020; 10:4208. PMID: 32144345.
Article48. Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011; 6:209–240. PMID: 21617937.
Article49. Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017; 2:135–143. PMID: 28403983.
Article50. Liu Z, Li N, Fang H, Chen X, Guo Y, Gong S, Niu M, Zhou H, Jiang Y, Chang P, Chen P. Enteric dysbiosis is associated with sepsis in patients. FASEB J. 2019; 33:12299–12310. PMID: 31465241.
Article51. Wan YD, Zhu RX, Wu ZQ, Lyu SY, Zhao LX, Du ZJ, Pan XT. Gut microbiota disruption in septic shock patients: a pilot study. Med Sci Monit. 2018; 24:8639–8646. PMID: 30488879.
Article52. Lankelma JM, van Vught LA, Belzer C, Schultz MJ, van der Poll T, de Vos WM, Wiersinga WJ. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2017; 43:59–68. PMID: 27837233.
Article53. Agudelo-Ochoa GM, Valdés-Duque BE, Giraldo-Giraldo NA, Jaillier-Ramírez AM, Giraldo-Villa A, Acevedo-Castaño I, Yepes-Molina MA, Barbosa-Barbosa J, Benítez-Paéz A. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes. 2020; 1–16.
Article54. Yin L, Wan YD, Pan XT, Zhou CY, Lin N, Ma CT, Yao J, Su Z, Wan C, Yu YW, Zhu RX. Association between gut bacterial diversity and mortality in septic shock patients: a cohort study. Med Sci Monit. 2019; 25:7376–7382. PMID: 31574078.
Article55. van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017; 17:407–420. PMID: 28436424.
Article56. Viaggi B, Poole D, Tujjar O, Marchiani S, Ognibene A, Finazzi S. Mid regional pro-adrenomedullin for the prediction of organ failure in infection. Results from a single centre study. PLoS One. 2018; 13:e0201491. PMID: 30102716.
Article57. Önal U, Valenzuela-Sánchez F, Vandana KE, Rello J. Mid-regional pro-adrenomedullin (MR-proADM) as a biomarker for sepsis and septic shock: narrative review. Healthcare (Basel). 2018; 6:E110. PMID: 30177659.
Article58. Christ-Crain M, Morgenthaler NG, Struck J, Harbarth S, Bergmann A, Müller B. Mid-regional pro-adrenomedullin as a prognostic marker in sepsis: an observational study. Crit Care. 2005; 9:R816–R824. PMID: 16356231.59. Andaluz-Ojeda D, Nguyen HB, Meunier-Beillard N, Cicuéndez R, Quenot JP, Calvo D, Dargent A, Zarca E, Andrés C, Nogales L, Eiros JM, Tamayo E, Gandía F, Bermejo-Martín JF, Charles PE. Superior accuracy of mid-regional proadrenomedullin for mortality prediction in sepsis with varying levels of illness severity. Ann Intensive Care. 2017; 7:15. PMID: 28185230.
Article60. Chow JW, Fine MJ, Shlaes DM, Quinn JP, Hooper DC, Johnson MP, Ramphal R, Wagener MM, Miyashiro DK, Yu VL. Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann Intern Med. 1991; 115:585–90. PMID: 1892329.
Article61. Pugin J, Auckenthaler R, Mili N, Janssens JP, Lew PD, Suter PM. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am Rev Respir Dis. 1991; 143:1121–1129. PMID: 2024824.
Article62. Weber J, Sachse J, Bergmann S, Sparwaßer A, Struck J, Bergmann A. Sandwich immunoassay for bioactive plasma adrenomedullin. J Appl Lab Med. 2017; 2:222–233.
Article63. Marino R, Struck J, Maisel AS, Magrini L, Bergmann A, Di Somma S. Plasma adrenomedullin is associated with short-term mortality and vasopressor requirement in patients admitted with sepsis. Crit Care. 2014; 18:R34. PMID: 24533868.
Article64. Kim H, Hur M, Struck J, Bergmann A, Di Somma S. Circulating biologically active adrenomedullin predicts organ failure and mortality in sepsis. Ann Lab Med. 2019; 39:454–463. PMID: 31037864.
Article65. Mebazaa A, Geven C, Hollinger A, Wittebole X, Chousterman BG, Blet A, Gayat E, Hartmann O, Scigalla P, Struck J, Bergmann A, Antonelli M, Beishuizen A, Constantin JM, Damoisel C, Deye N, Di Somma S, Dugernier T, François B, Gaudry S, Huberlant V, Lascarrou JB, Marx G, Mercier E, Oueslati H, Pickkers P, Sonneville R, Legrand M, Laterre PF. AdrenOSS-1 study investigators. Circulating adrenomedullin estimates survival and reversibility of organ failure in sepsis: the prospective observational multinational Adrenomedullin and Outcome in Sepsis and Septic Shock-1 (AdrenOSS-1) study. Crit Care. 2018; 22:354. PMID: 30583748.
Article66. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009; 11:228–234. PMID: 19255566.
Article67. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010; 11:597–610. PMID: 20661255.
Article68. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Voinea SC. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020; 9:E276. PMID: 31979244.
Article69. Szilágyi B, Fejes Z, Pócsi M, Kappelmayer J, Nagy B Jr. Role of sepsis modulated circulating microRNAs. EJIFCC. 2019; 30:128–145. PMID: 31263389.70. Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, Nicoloso MS, Barbarotto E, Popa M, Stanciulea O, Fernandez MH, Tulbure D, Bueso-Ramos CE, Negrini M, Calin GA. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009; 4:e7405. PMID: 19823581.
Article71. Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS One. 2012; 7:e38885. PMID: 22719975.
Article72. Li H, Ding G. Elevated serum inflammatory cytokines in lupus nephritis patients, in association with promoted hsa-miR-125a. Clin Lab. 2016; 62:631–638. PMID: 27215082.
Article73. Hu HL, Nie ZQ, Lu Y, Yang X, Song C, Chen H, Zhu S, Chen BB, Huang J, Geng S, Zhao S. Circulating miR-125b but not miR-125a correlates with acute exacerbations of chronic obstructive pulmonary disease and the expressions of inflammatory cytokines. Medicine (Baltimore). 2017; 96:e9059. PMID: 29390434.
Article74. Zhao D, Li S, Cui J, Wang L, Ma X, Li Y. Plasma miR-125a and miR-125b in sepsis: Correlation with disease risk, inflammation, severity, and prognosis. J Clin Lab Anal. 2020; 34:e23036. PMID: 32077163.
Article75. Hermans-Beijnsberger S, van Bilsen M, Schroen B. Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res. 2018; 3:118–130. PMID: 30175285.
Article76. Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014; 35:408–419. PMID: 25113636.
Article77. Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018; 70:1038–1053.e7. PMID: 29932899.
Article78. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014; 53:393–406. PMID: 24507715.
Article79. He F, Zhang C, Huang Q. Long noncoding RNA nuclear enriched abundant transcript 1/miRNA-124 axis correlates with increased disease risk, elevated inflammation, deteriorative disease condition, and predicts decreased survival of sepsis. Medicine (Baltimore). 2019; 98:e16470. PMID: 31393351.
Article80. Huang Q, Huang C, Luo Y, He F, Zhang R. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med. 2018; 36:1659–1663. PMID: 29936011.
Article81. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017; 14:1705–1714. PMID: 28837398.
Article82. Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016; 590:2884–2895. PMID: 27434861.
Article83. Chen J, He Y, Zhou L, Deng Y, Si L. Long non-coding RNA MALAT1 serves as an independent predictive biomarker for the diagnosis, severity and prognosis of patients with sepsis. Mol Med Rep. 2020; 21:1365–1373. PMID: 31922243.
Article84. Na L, Ding H, Xing E, Gao J, Liu B, Wang H, Yu J, Yu C. Lnc-MEG3 acts as a potential biomarker for predicting increased disease risk, systemic inflammation, disease severity, and poor prognosis of sepsis via interacting with miR-21. J Clin Lab Anal. 2020; e23123. PMID: 31907972.
Article85. Zha F, Qu X, Tang B, Li J, Wang Y, Zheng P, Ji T, Zhu C, Bai S. Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 2019; 11:3716–3730. PMID: 31195367.
Article86. Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys. 2019; 661:22–30. PMID: 30389444.
Article87. Parikh SM. The Angiopoietin-Tie2 Signaling Axis in Systemic Inflammation. J Am Soc Nephrol. 2017; 28:1973–1982. PMID: 28465380.
Article88. Leligdowicz A, Richard-Greenblatt M, Wright J, Crowley VM, Kain KC. Endothelial activation: The Ang/Tie axis in sepsis. Front Immunol. 2018; 9:838. PMID: 29740443.
Article89. Saharinen P, Eklund L, Alitalo K. Therapeutic targeting of the angiopoietin-TIE pathway. Nat Rev Drug Discov. 2017; 16:635–661. PMID: 28529319.
Article90. Seol CH, Yong SH, Shin JH, Lee SH, Leem AY, Park SM, Kim YS, Chung KS. The ratio of plasma angiopoietin-2 to angiopoietin-1 as a prognostic biomarker in patients with sepsis. Cytokine. 2020; 129:155029. PMID: 32059166.
Article91. Fang Y, Li C, Shao R, Yu H, Zhang Q, Zhao L. Prognostic significance of the angiopoietin-2/angiopoietin-1 and angiopoietin-1/Tie-2 ratios for early sepsis in an emergency department. Crit Care. 2015; 19:367. PMID: 26463042.
Article92. Zhang Y, Gan C, Zhang J, Chen D. LPS-induced downregulation of microRNA-204/211 upregulates and stabilizes Angiopoietin-1 mRNA in EA.hy926 endothelial cells. Mol Med Rep. 2017; 16:6081–6087. PMID: 28901393.
Article93. ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992; 20:864–874. PMID: 1597042.94. Aksaray S, Alagoz P, Inan A, Cevan S, Ozgultekin A. Diagnostic value of sTREM-1 and procalcitonin levels in the early diagnosis of sepsis. North Clin Istanb. 2016; 3:175–182. PMID: 28275748.
Article95. Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. SCCM/ESICM/ACCP/ATS/SIS. SCCM/ESICM/ACCP/ATS/SIS. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003; 31:1250–1256. PMID: 12682500.96. Brenner T, Uhle F, Fleming T, Wieland M, Schmoch T, Schmitt F, Schmidt K, Zivkovic AR, Bruckner T, Weigand MA, Hofer S. Soluble TREM-1 as a diagnostic and prognostic biomarker in patients with septic shock: an observational clinical study. Biomarkers. 2017; 22:63–69. PMID: 27319606.
Article97. Spoto S, Cella E, de Cesaris M, Locorriere L, Mazzaroppi S, Nobile E, Lanotte AM, Pedicino L, Fogolari M, Costantino S, Dicuonzo G, Ciccozzi M, Angeletti S. Procalcitonin and MR-proadrenomedullin combination with SOFA and qSOFA scores for sepsis diagnosis and prognosis: a diagnostic algorithm. Shock. 2018; 50:44–52. PMID: 29023361.
Article98. Casagranda I, Vendramin C, Callegari T, Vidali M, Calabresi A, Ferrandu G, Cervellin G, Cavazza M, Lippi G, Zanotti I, Negro S, Rocchetti A, Arfini C. Usefulness of suPAR in the risk stratification of patients with sepsis admitted to the emergency department. Intern Emerg Med. 2015; 10:725–730. PMID: 26156446.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Eosinophil count and neutrophil-to-lymphocyte count ratio as biomarkers for predicting early-onset neonatal sepsis
- Biomarkers of Sepsis
- Sepsis and Acute Respiratory Distress Syndrome: Recent Update
- Infectious diseases and biomarker use
- Difference in Protein Markers According to the Survival of Sepsis Patients using Protein Chips