J Vet Sci.
2020 Mar;21(2):e20. 10.4142/jvs.2020.21.e20.
Construction and immunization with double mutant ΔapxIBD Δpnp forms of Oryctolagus cuniculus serotypes 1 and 5
- Affiliations
-
- 1College of Veterinary Medicine and Institute of Veterinary Science, Kangwon National University, Chuncheon 24341, Korea
- 2Key Laboratory of Veterinary Medicine, Faculty of Veterinary Medicine, Vietnam National University of Agriculture, Hanoi 100000, Vietnam
- KMID: 2506179
- DOI: http://doi.org/10.4142/jvs.2020.21.e20
Abstract
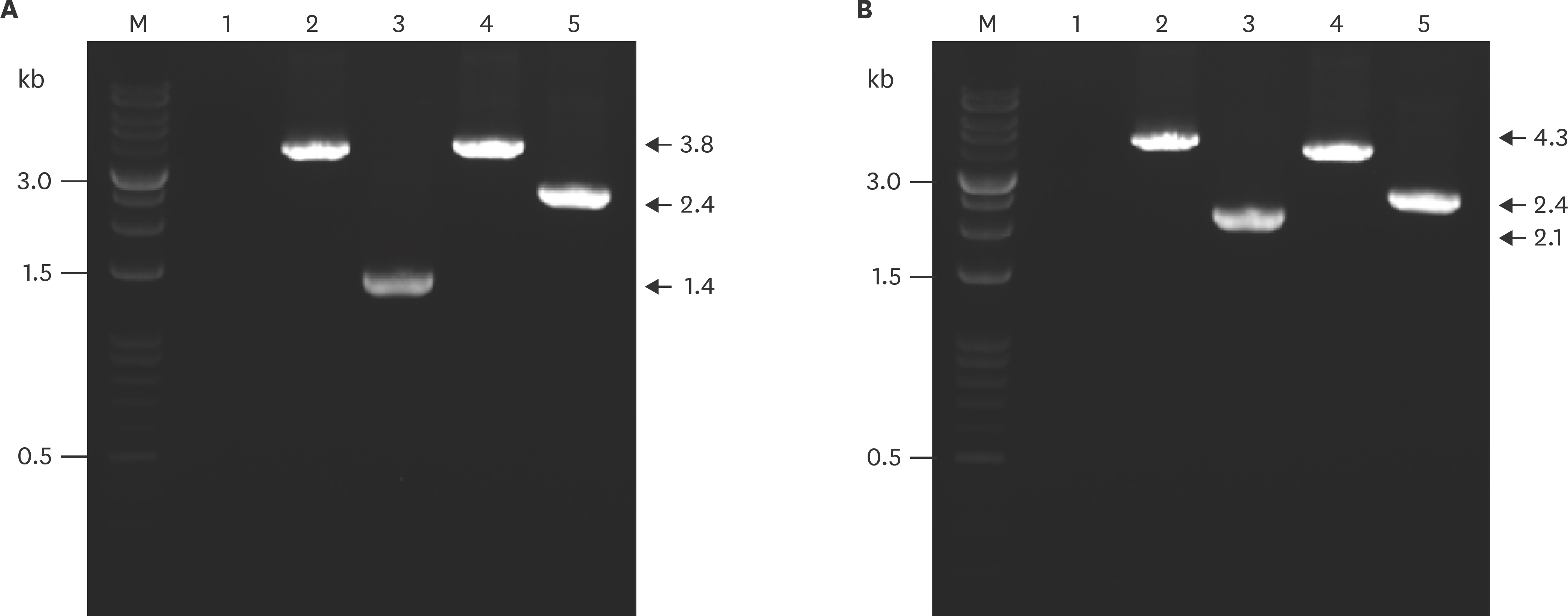
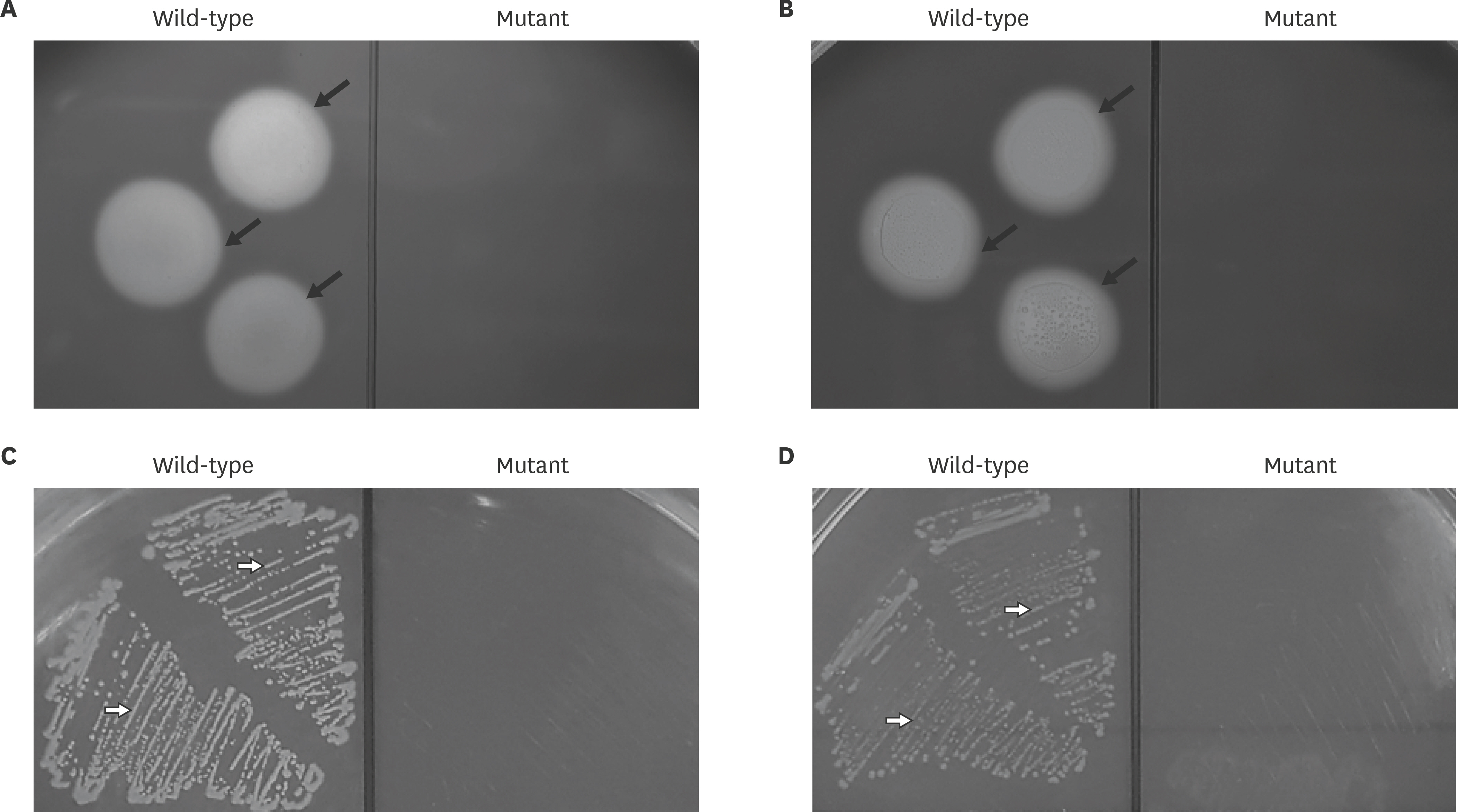
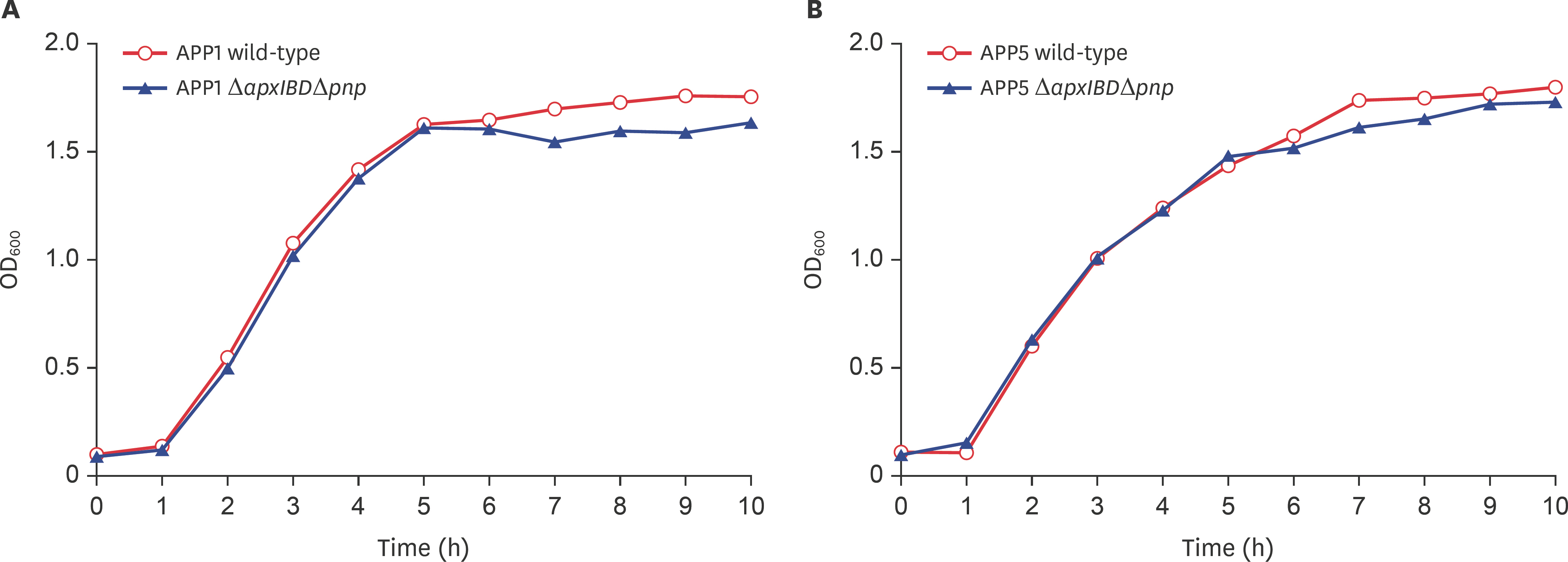
- Actinobacillus pleuropneumoniae (APP) causes a form of porcine pleuropneumonia that leads to significant economic losses in the swine industry worldwide. The apxIBD gene is responsible for the secretion of the ApxI and ApxII toxins and the pnp gene is responsible for the adaptation of bacteria to cold temperature and a virulence factor. The apxIBDand pnp genes were deleted successfully from APP serotype 1 and 5 by transconjugation and sucrose counter-selection. The APP1ΔapxIBD Δpnp and APP5ΔapxIBD Δpnp mutants lost hemolytic activity and could not secrete ApxI and ApxII toxins outside the bacteria because both mutants lost the ApxI- and ApxII-secreting proteins by deletion of the apxIBD gene. Besides, the growth of these mutants was defective at low temperatures resulting from the deletion of pnp. The APP1ΔapxIBD Δpnp and APP5ΔapxIBD Δpnp mutants were significantly attenuated compared with wild-type ones. However, mice vaccinated intraperitoneally with APP5ΔapxIBD Δpnpdid not provide any protection when challenged with a 10-times 50% lethal dose of virulent homologous (APP5) and heterologous (APP1) bacterial strains, while mice vaccinated with APP1ΔapxIBD Δpnp offered 75% protection against a homologous challenge. The ΔapxIBD Δpnp mutants were significantly attenuated and gave different protection rate against homologous virulent wild-type APP challenging.
Figure
Reference
-
References
1. Sassu EL, Bossé JT, Tobias TJ, Gottschalk M, Langford PR, Hennig-Pauka I. Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound Emerg Dis. 2018; 65(Suppl 1):72–90.2. Bossé JT, Li Y, Sárközi R, Fodor L, Lacouture S, Gottschalk M, Casas Amoribieta M, Angen Ø, Nedbalcova K, Holden MT, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR. BRaDP1T consortium. Proposal of serovars 17 and 18 of Actinobacillus pleuropneumoniae based on serological and genotypic analysis. Vet Microbiol. 2018; 217:1–6.3. Lee KE, Choi HW, Kim HH, Song JY, Yang DK. Prevalence and characterization of Actinobacillus pleuropneumoniae isolated from Korean pigs. J Bacteriol Virol. 2015; 45:19–25.4. Komal JP, Mittal KR. Grouping of Actinobacillus pleuropneumoniae strains of serotypes 1 through 12 on the basis of their virulence in mice. Vet Microbiol. 1990; 25:229–240.5. Yuan F, Liu J, Guo Y, Tan C, Fu S, Zhao J, Chen H, Bei W. Influences of ORF1 on the virulence and immunogenicity of Actinobacillus pleuropneumoniae. Curr Microbiol. 2011; 63:574–580.6. Frey J. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 1995; 3:257–261.7. Jansen R, Briaire J, Kamp EM, Gielkens AL, Smits MA. Structural analysis of the Actinobacillus pleuropneumoniae-RTX-toxin I (ApxI) operon. Infect Immun. 1993; 61:3688–3695.8. Jones GH. Novel aspects of polynucleotide phosphorylase function in Streptomyces. Antibiotics (Basel). 2018; 7:25.9. Barria C, Malecki M, Arraiano CM. Bacterial adaptation to cold. Microbiology. 2013; 159:2437–2443.
Article10. Rosenzweig JA, Chopra AK. The exoribonuclease polynucleotide phosphorylase influences the virulence and stress responses of yersiniae and many other pathogens. Front Cell Infect Microbiol. 2013; 3:81.
Article11. De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011; 17:1172–1189.12. Cameron TA, Matz LM, De Lay NR. Polynucleotide phosphorylase: not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 2018; 14:e1007654.
Article13. Fuller TE, Kennedy MJ, Lowery DE. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb Pathog. 2000; 29:25–38.14. Engman J, Negrea A, Sigurlásdóttir S, Geörg M, Eriksson J, Eriksson OS, Kuwae A, Sjölinder H, Jonsson AB. Neisseria meningitidis polynucleotide phosphorylase affects aggregation, adhesion, and virulence. Infect Immun. 2016; 84:1501–1513.15. Fuller TE, Martin S, Teel JF, Alaniz GR, Kennedy MJ, Lowery DE. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb Pathog. 2000; 29:39–51.16. Oswald W, Tonpitak W, Ohrt G, Gerlach G. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol Lett. 1999; 179:153–160.17. Lin L, Bei W, Sha Y, Liu J, Guo Y, Liu W, Tu S, He Q, Chen H. Construction and immunogencity of a ΔapxIC/ΔapxIIC double mutant of Actinobacillus pleuropneumoniae serovar 1. FEMS Microbiol Lett. 2007; 274:55–62.18. Hu J, McCormick RJ, Means WJ, Zhu MJ. Polynucleotide phosphorylase is required for Escherichia coli O157: H7 growth above refrigerated temperature. Foodborne Pathog Dis. 2014; 11:177–185.19. Nielsen R, van den Bosch JF, Plambeck T, Sørensen V, Nielsen JP. Evaluation of an indirect enzyme-linked immunosorbent assay (ELISA) for detection of antibodies to the Apx toxins of Actinobacillus pleuropneumoniae. Vet Microbiol. 2000; 71:81–87.20. Xu F, Chen X, Shi A, Yang B, Wang J, Li Y, Guo X, Blackall PJ, Yang H. Characterization and immunogenicity of an apxIA mutant of Actinobacillus pleuropneumoniae. Vet Microbiol. 2006; 118:230–239.21. Bei W, He Q, Yan L, Fang L, Tan Y, Xiao S, Zhou R, Jin M, Guo A, Lv J, Huang H, Chen H. Construction and characterization of a live, attenuated apxIICA inactivation mutant of Actinobacillus pleuropneumoniae lacking a drug resistance marker. FEMS Microbiol Lett. 2005; 243:21–27.22. Liu J, Chen X, Lin L, Tan C, Chen Y, Guo Y, Jin M, Guo A, Bei W, Chen H. Potential use an Actinobacillus pleuropneumoniae double mutant strain ΔapxIICΔapxIVA as live vaccine that allows serological differentiation between vaccinated and infected animals. Vaccine. 2007; 25:7696–7705.23. Seah JN, Frey J, Kwang J. The N-terminal domain of RTX toxin ApxI of Actinobacillus pleuropneumoniae elicits protective immunity in mice. Infect Immun. 2002; 70:6464–6467.24. Frey J. The role of RTX toxins in host specificity of animal pathogenic Pasteurellaceae. Vet Microbiol. 2011; 153:51–58.25. Ramjeet M, Deslandes V, Gouré J, Jacques M. Actinobacillus pleuropneumoniae vaccines: from bacterins to new insights into vaccination strategies. Anim Health Res Rev. 2008; 9:25–45.26. Yuan F, Liao Y, You W, Liu Z, Tan Y, Zheng C, Wang Bin, Zhou D, Tian Y, Bei W. Deletion of the znuA virulence factor attenuates Actinobacillus pleuropneumoniae and confers protection against homologous or heterologous strain challenge. Vet Microbiol. 2014; 174:531–539.27. Xie F, Li G, Zhou L, Zhang Y, Cui N, Liu S, Wang C. Attenuated Actinobacillus pleuropneumoniae double-deletion mutant S-8∆clpP/apxIIC confers protection against homologous or heterologous strain challenge. BMC Vet Res. 2017; 13:14.
Article28. Wang L, Qin W, Yang S, Zhai R, Zhou L, Sun C, Pan F, Ji Q, Wang Y, Gu J, Feng X, Du C, Han W, Langford PR, Lei L. The Adh adhesin domain is required for trimeric autotransporter Apa1-mediated Actinobacillus pleuropneumoniae adhesion, autoaggregation, biofilm formation and pathogenicity. Vet Microbiol. 2015; 177:175–183.29. Fu S, Ou J, Zhang M, Xu J, Liu H, Liu J, Yuan F, Chen H, Bei W. The live attenuated Actinobacillus pleuropneumoniae triple-deletion mutant ΔapxIC ΔapxIIC ΔapxIV-ORF1 strain, SLW05, Immunizes pigs against lethal challenge with Haemophilus parasuis. Clin Vaccine Immunol. 2013; 20:134–139.30. Chiers K, De Waele T, Pasmans F, Ducatelle R, Haesebrouck F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet Res. 2010; 41:65.31. Park C, Ha Y, Kim S, Chae C, Ryu DY. Construction and characterization of an Actinobacillus pleuropneumoniae serotype 2 mutant lacking the Apx toxin secretion protein genes apxIIIB and apxIIID. J Vet Med Sci. 2009; 71:1317–1323.32. Bao CT, Xiao JM, Liu BJ, Liu JF, Zhu RN, Jiang P, Li L, Langford PR, Lei LC. Establishment and comparison of Actinobacillus pleuropneumoniae experimental infection model in mice and piglets. Microb Pathog. 2019; 128:381–389.33. Li Y, Cao S, Zhang L, Yuan J, Zhao Q, Wen Y, Wu R, Huang X, Yan Q, Huang Y, Ma X, Han X, Miao C, Wen X. A requirement of TolC1 for effective survival, colonization and pathogenicity of Actinobacillus pleuropneumoniae. Microb Pathog. 2019; 134:103596.34. Li T, Zhang Q, Wang R, Zhang S, Pei J, Li Y, Li L, Zhou R. The roles of flp1 and tadD in Actinobacillus pleuropneumoniae pilus biosynthesis and pathogenicity. Microb Pathog. 2019; 126:310–317.35. Loera-Muro A, Angulo C. New trends in innovative vaccine development against Actinobacillus pleuropneumoniae. Vet Microbiol. 2018; 217:66–75.36. Inzana TJ, Todd J, Ma JN, Veit H. Characterization of a non-hemolytic mutant of Actinobacillus pleuropneumoniae serotype 5: role of the 110 kilodalton hemolysin in virulence and immunoprotection. Microb Pathog. 1991; 10:281–296.37. Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997; 179:538–540.38. Baltes N, Tonpitak W, Hennig-Pauka I, Gruber AD, Gerlach GF. Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol Lett. 2003; 220:41–48.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Inactivated pep27 mutant as an effective mucosal vaccine against a secondary lethal pneumococcal challenge in mice
- Functional Antibody Responses to Seven Serotypes in Pneumococcal Polysaccharide Vaccine in Children
- Seroprevalence of Encephalitozoon cuniculi and Toxoplasma gondii in domestic rabbits (Oryctolagus cuniculus) in China
- Cross-reaction of 6B and 19F Specific Antibodies to Serotypes 6A, 6C, and 19A after Immunization with 7-valent Pneumococcal Conjugate Vaccine in Korean Children Aged 12-23 Months
- Distribution of Hepatitis C Virus Serotypes of HCV RNA Positive Subjects in Korea