Nutr Res Pract.
2020 Aug;14(4):299-308. 10.4162/nrp.2020.14.4.299.
Effect of probiotics Lactobacillus paracasei GKS6, L. plantarum GKM3, and L. rhamnosus GKLC1 on alleviating alcohol-induced alcoholic liver disease in a mouse model
- Affiliations
-
- 1Biotech Research Institute, Grape King Bio Ltd, Taoyuan 32542, Taiwan
- 2Institute of Food Science and Technology, National Taiwan University, Taipei 10617, Taiwan
- 3Department of Food Science, Nutrition and Nutraceutical Biotechnology, Shih Chien University, Taipei 10462, Taiwan
- 4Department of Bioscience Technology, Chung Yuan Christian University, Taoyuan 32023, Taiwan
- KMID: 2504747
- DOI: http://doi.org/10.4162/nrp.2020.14.4.299
Abstract
- BACKGROUND/OBJECTIVES
Heavy alcohol consumption causes the development of alcoholic liver disease (ALD), a neglected but important public health problem. Many studies have pointed out that probiotics could improve gut health, which is also considered to be a cause of ALD. Therefore, this study screened the probiotics, Lactobacillus casei GKC1 (GKC1), L. fermentum GKF3 (GKF3), Bifidobacterium lactis GKK2 (GKK2), L. rhamnosus GKLC1 (GKLC1), L. paracasei GKS6 (GKS6), and L. plantarum GKM3 (GKM3), for their potential benefits in alleviating ALD for applications to disease prevention.
SUBJECTS/METHODS
C57BL/6N mice were divided into 8 groups (n = 6 in each): normal control, positive control (alcohol-diet fed), and treatments of feeding probiotics GKC1, GKF3, GKK2, GKLC1, GKS6, and GKM3 under an oral dose 0.82 g/kg B.W. per day by oral gavage. The experiment was conducted for 8 weeks, and the concentrations of alanine aminotransferase (ALT), aspartate aminotransferase, triglyceride (TG), and total cholesterol (TC) in mice were measured. The glutathione (GSH), catalase (CAT), and histology were analyzed after sacrifice.
RESULTS
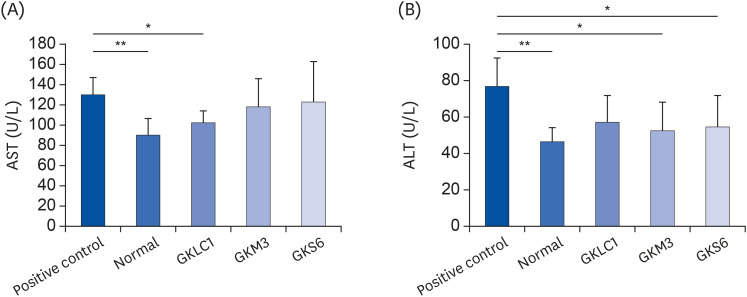
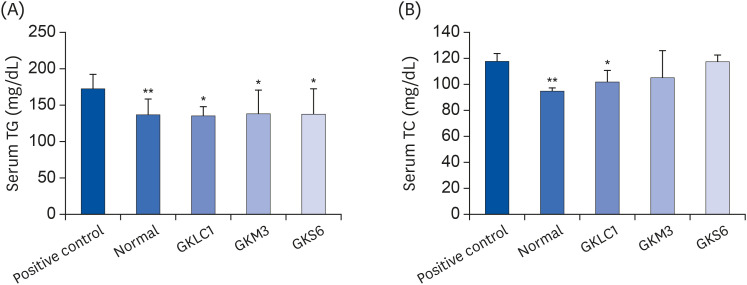
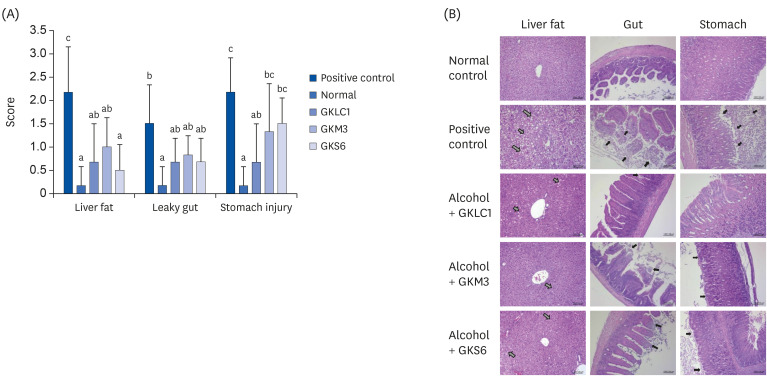
The results showed a decrease in the serum ALT, liver TG, and liver TC levels in the GKS6, GKM3, and GKLC1 groups compared to the positive control. In addition, the decreasing GSH and CAT levels were inhibited in the GKS6 and GKM3 groups. The histopathological results showed that all probiotics could reduce the accumulation of liver fat. Furthermore, there was a significant difference in GKLC1 with lower stomach damage compared to the alcohol-fed mice without any addition of probiotics.
CONCLUSIONS
GKLC1, GKS6, and GKM3 can be used as supplements for alleviating the development of ALD.
Figure
Reference
-
1. Dey A, Cederbaum AI. Alcohol and oxidative liver injury. Hepatology. 2006; 43:S63–S74. PMID: 16447273.
Article2. Nagata K, Suzuki H, Sakaguchi S. Common pathogenic mechanism in development progression of liver injury caused by non-alcoholic or alcoholic steatohepatitis. J Toxicol Sci. 2007; 32:453–468. PMID: 18198478.
Article3. Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J Gastroenterol. 2014; 20:17756–17772. PMID: 25548474.
Article4. Ringseis R, Muschick A, Eder K. Dietary oxidized fat prevents ethanol-induced triacylglycerol accumulation and increases expression of PPARalpha target genes in rat liver. J Nutr. 2007; 137:77–83. PMID: 17182804.5. Masella R, Di Benedetto R, Varì R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005; 16:577–586. PMID: 16111877.
Article6. Nanji AA. Role of Kupffer cells in alcoholic hepatitis. Alcohol. 2002; 27:13–15. PMID: 12062631.
Article7. Wheeler MD. Endotoxin and Kupffer cell activation in alcoholic liver disease. Alcohol Res Health. 2003; 27:300–306. PMID: 15540801.8. Cesaro C, Tiso A, Del Prete A, Cariello R, Tuccillo C, Cotticelli G, Del Vecchio Blanco C, Loguercio C. Gut microbiota and probiotics in chronic liver diseases. Dig Liver Dis. 2011; 43:431–438. PMID: 21163715.
Article9. Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011; 141:1572–1585. PMID: 21920463.
Article10. Nanji AA, Khettry U, Sadrzadeh SM. Lactobacillus feeding reduces endotoxemia and severity of experimental alcoholic liver (disease). Proc Soc Exp Biol Med. 1994; 205:243–247. PMID: 8171045.11. Wang Y, Kirpich I, Liu Y, Ma Z, Barve S, McClain CJ, Feng W. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am J Pathol. 2011; 179:2866–2875. PMID: 22093263.12. Kirpich IA, Solovieva NV, Leikhter SN, Shidakova NA, Lebedeva OV, Sidorov PI, Bazhukova TA, Soloviev AG, Barve SS, McClain CJ, Cave M. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008; 42:675–682. PMID: 19038698.
Article13. Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010; 7:503–514. PMID: 20664519.
Article14. Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013; 6:39–51.
Article15. Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013; 8:e53028. PMID: 23326376.16. Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 2009; 43:163–172. PMID: 19251117.17. Wang Y, Liu Y, Sidhu A, Ma Z, McClain C, Feng W. Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am J Physiol Gastrointest Liver Physiol. 2012; 303:G32–41. PMID: 22538402.18. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957; 226:497–509. PMID: 13428781.
Article19. Ryu JE, Jo W, Choi HJ, Jang S, Lee HJ, Woo DC, Kim JK, Kim KW, Yu ES, Son WC. Evaluation of nonalcoholic fatty liver disease in C57BL/6J mice by using MRI and histopathologic analyses. Comp Med. 2015; 65:409–415. PMID: 26473344.20. Shackelford C, Long G, Wolf J, Okerberg C, Herbert R. Qualitative and quantitative analysis of nonneoplastic lesions in toxicology studies. Toxicol Pathol. 2002; 30:93–96. PMID: 11890482.
Article21. Osna NA, Donohue TM Jr, Kharbanda KK. Alcoholic liver disease: pathogenesis and current management. Alcohol Res. 2017; 38:147–161. PMID: 28988570.22. Ahmed M. Non-alcoholic fatty liver disease in 2015. World J Hepatol. 2015; 7:1450–1459. PMID: 26085906.
Article23. Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002; 109:1125–1131. PMID: 11994399.
Article24. Clugston RD, Jiang H, Lee MX, Piantedosi R, Yuen JJ, Ramakrishnan R, Lewis MJ, Gottesman ME, Huang LS, Goldberg IJ, Berk PD, Blaner WS. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J Lipid Res. 2011; 52:2021–2031. PMID: 21856784.
Article25. Sun JR. Antiobesity effects of lactic acid bacteria supplements on high-fat diet-induced obese rats and their molecular mechanisms [master's thesis]. Taichung: Chung Shan Medical University;2017.26. Salvaggio A, Periti M, Miano L, Tavanelli M, Marzorati D. Body mass index and liver enzyme activity in serum. Clin Chem. 1991; 37:720–723. PMID: 1674452.
Article27. Ewaschuk J, Endersby R, Thiel D, Diaz H, Backer J, Ma M, Churchill T, Madsen K. Probiotic bacteria prevent hepatic damage and maintain colonic barrier function in a mouse model of sepsis. Hepatology. 2007; 46:841–850. PMID: 17659579.
Article28. Gutiérrez-Ruiz MC, Quiroz SC, Souza V, Bucio L, Hernández E, Olivares IP, Llorente L, Vargas-Vorácková F, Kershenobich D. Cytokines, growth factors, and oxidative stress in HepG2 cells treated with ethanol, acetaldehyde, and LPS. Toxicology. 1999; 134:197–207. PMID: 10403637.
Article29. Clark VL, Kruse JA. Clinical methods: the history, physical, and laboratory examinations. JAMA. 1990; 264:2808–2809.
Article30. Giannini EG, Testa R, Savarino V. Liver enzyme alteration: a guide for clinicians. CMAJ. 2005; 172:367–379. PMID: 15684121.
Article31. Segawa S, Wakita Y, Hirata H, Watari J. Oral administration of heat-killed Lactobacillus brevis SBC8803 ameliorates alcoholic liver disease in ethanol-containing diet-fed C57BL/6N mice. Int J Food Microbiol. 2008; 128:371–377. PMID: 18976829.32. Rao R. Endotoxemia and gut barrier dysfunction in alcoholic liver disease. Hepatology. 2009; 50:638–644. PMID: 19575462.
Article33. Patel D, Patel F, Mandal P. Potential molecular mechanism of probiotics in alcoholic liver disease. J Alcohol Drug Depend. 2017; 5:278.
Article34. Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 2010; 298:G807–19. PMID: 20299599.
Article35. Chen X, Zhang J, Yi R, Mu J, Zhao X, Yang Z. Hepatoprotective effects of Lactobacillus on carbon tetrachloride-induced acute liver injury in mice. Int J Mol Sci. 2018; 19:2212.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- High-quality draft genome and characterization of commercially potent probiotic Lactobacillus strains
- The effects of three novel probiotics isolated from the Korean fermented food Kimchi on the stress-induced defecation of rats
- Beneficial Effects of Lactobacillus casei ATCC 334 on Halitosis Induced by Periodontopathogens
- The Effects of Lactobacillus rhamnosus on the Prevention of Asthma in a Murine Model
- Efficacy of Probiotic Therapy on Atopic Dermatitis in Children: A Randomized, Double-blind, Placebo-controlled Trial