Cancer Res Treat.
2020 Jul;52(3):779-788. 10.4143/crt.2019.700.
Clinical Implications of Circulating Tumor DNA from Ascites and Serial Plasma in Ovarian Cancer
- Affiliations
-
- 1Division of Life Sciences, College of Life Sciences and Bioengineering, Incheon National University, Incheon, Korea
- 2Department of Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Cancer Evolution Research Center, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 4Department of Obstetrics and Gynecology, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 5Department of Laboratory Medicine, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea
- 6Cancer Research Institute, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2504459
- DOI: http://doi.org/10.4143/crt.2019.700
Abstract
- Purpose
The purpose of this study was to identify the clinical utility of circulating tumor DNA (ctDNA) from ascites and serial plasma samples from epithelial ovarian cancer (EOC) patients.
Materials and Methods
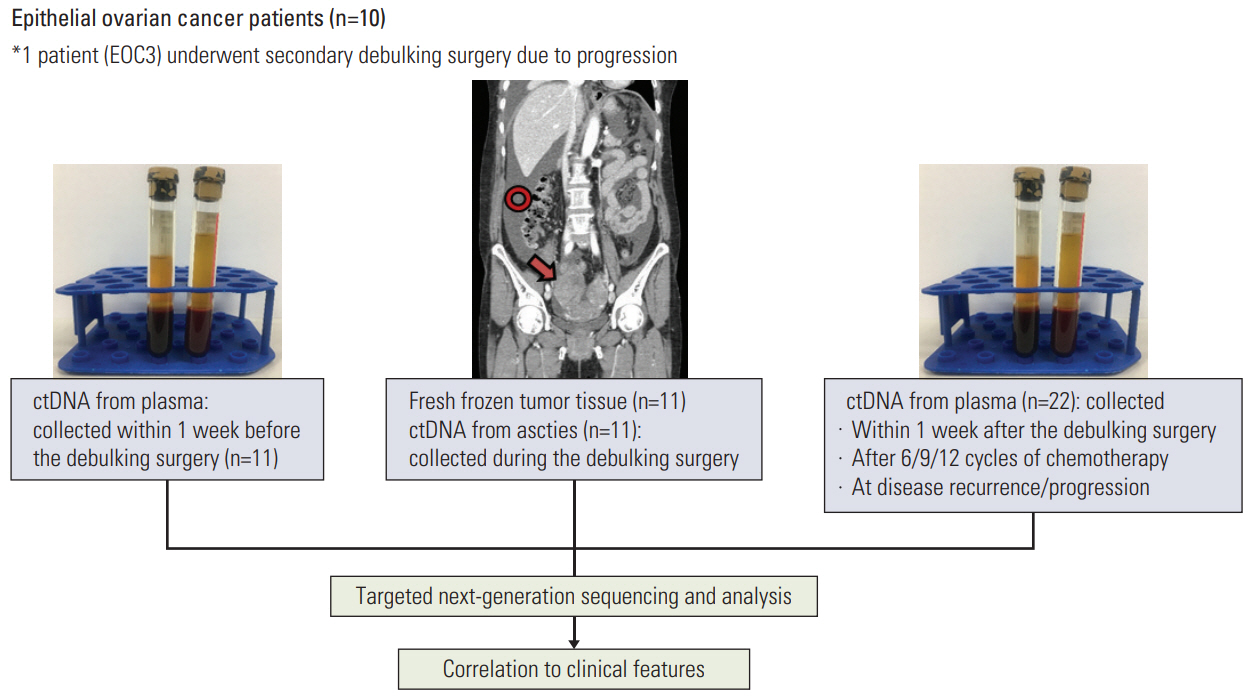
Using targeted next-generation sequencing, we analyzed a total of 55 EOC samples including ctDNA from ascites and serial plasma and gDNA from tumor tissues. Tumor tissues and ascites were collected during debulking surgeries and plasma samples were collected before and after the surgeries. Because one EOC patient underwent secondary debulking surgery, a total of 11 tumor tissues, 33 plasma samples, and 11 ascites samples were obtained from the 10 patients.
Results
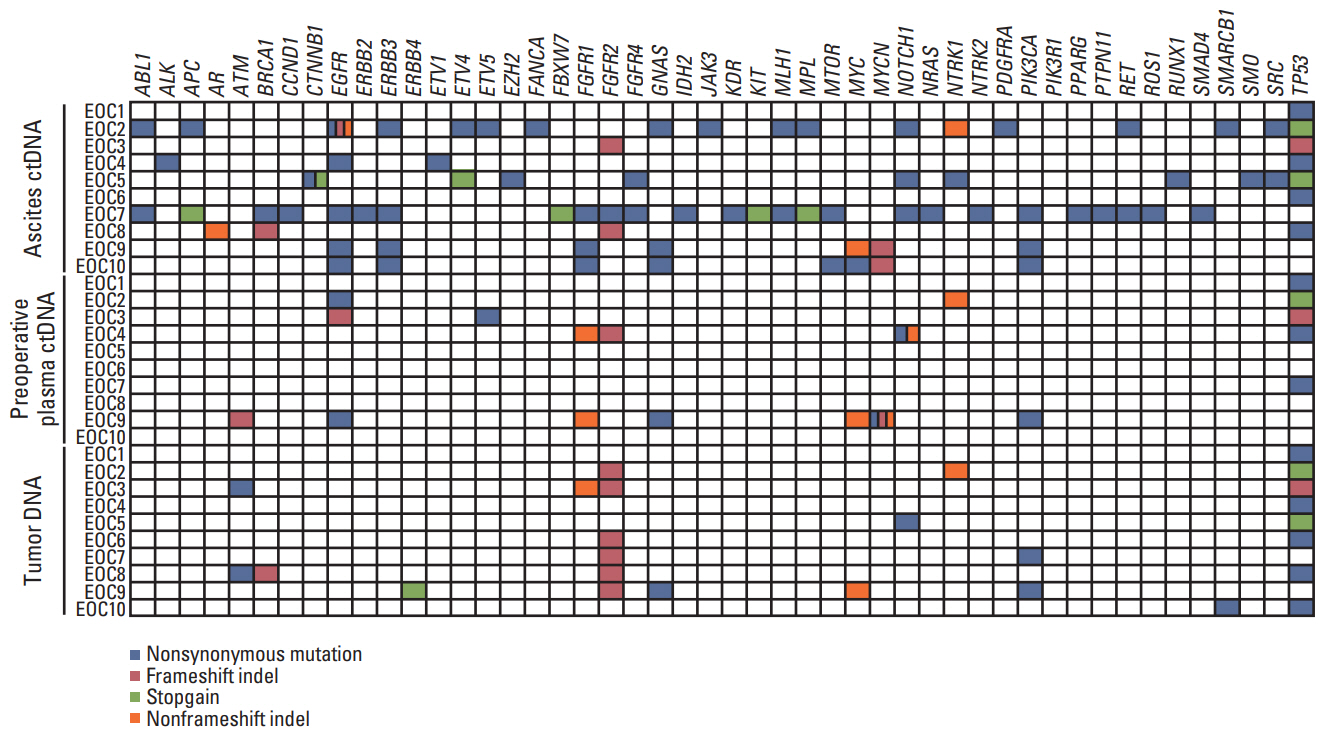
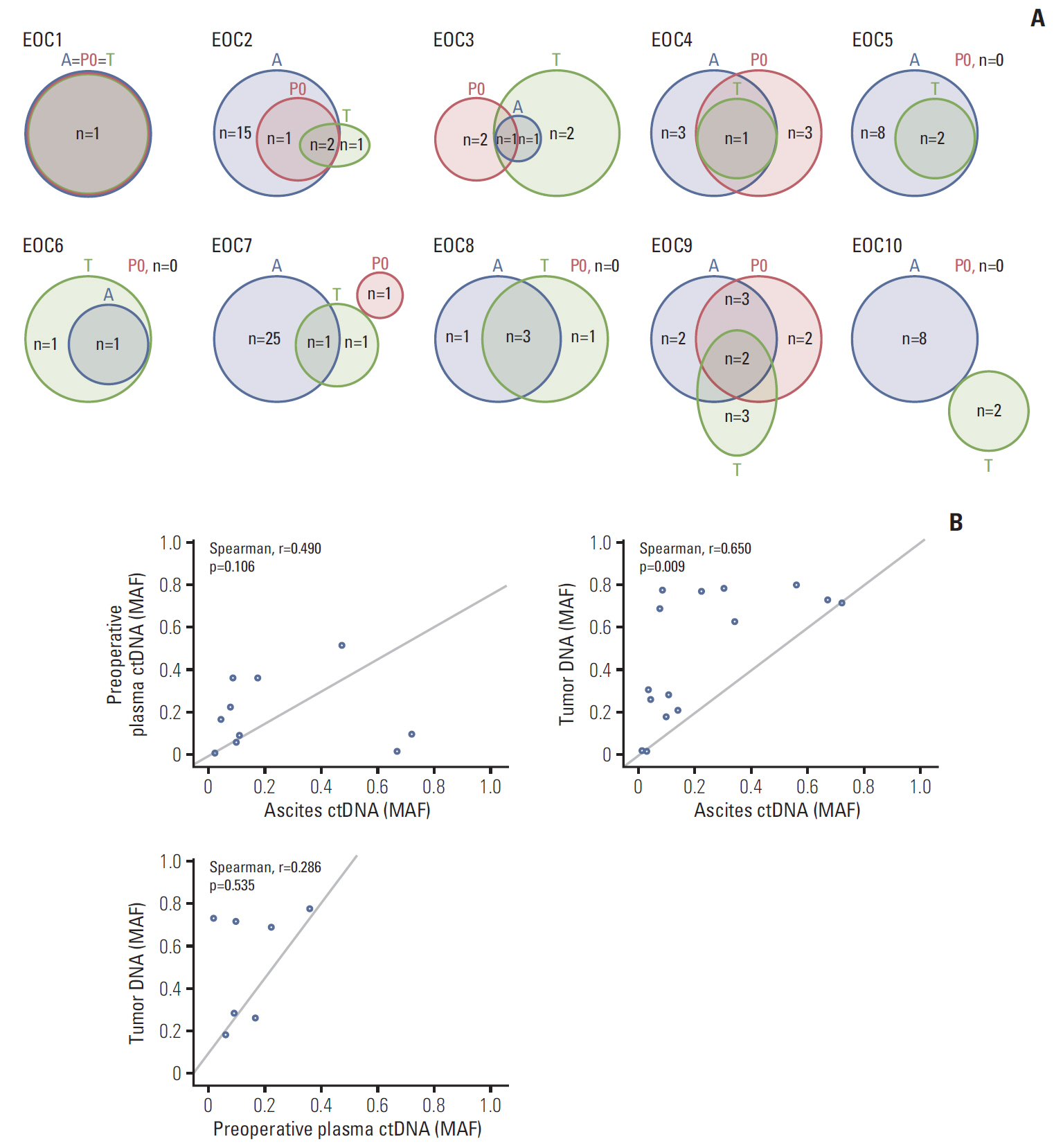
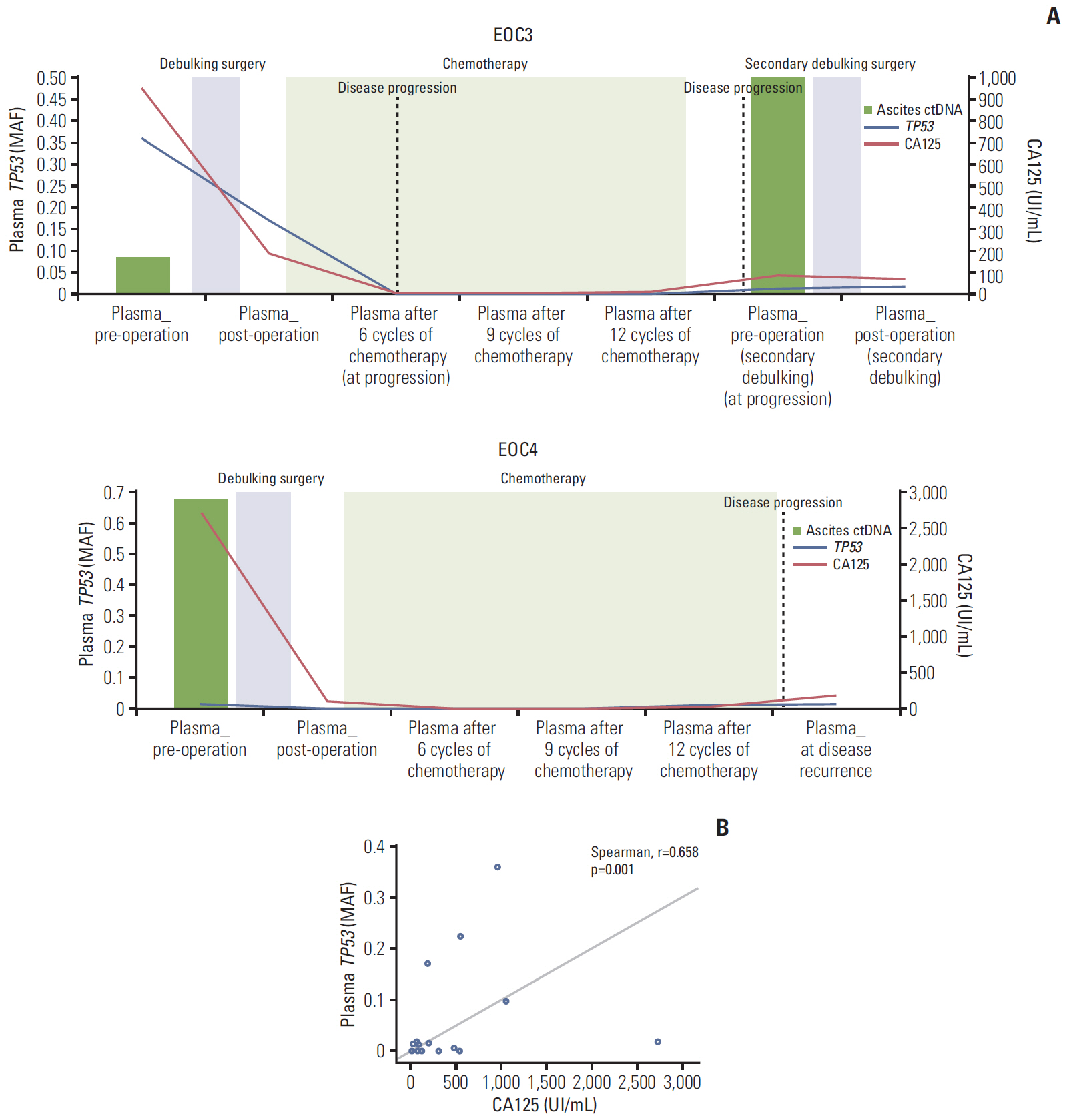
Of the 10 patients, nine (90%) contained somatic mutations in both tumor tissues and ascites ctDNA. This mutational concordance was confirmed through correlation analysis. The mutational concordance between ascites and tumor tissues was valid in recurrent/progressive ovarian cancer. TP53 was the most frequently detected gene with mutations. ctDNA from serial plasma samples identified EOC progression/recurrence at a similar time or even more rapidly than cancer antigen 125, an established serum protein tumor marker for EOC.
Conclusion
Our data suggest that ascites ctDNA can be used to identify the mutational landscape of ovarian cancer for therapeutic strategy planning.
Figure
Reference
-
References
1. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018; 68:284–96.
Article2. Clarke-Pearson DL. Clinical practice. Screening for ovarian cancer. N Engl J Med. 2009; 361:170–7.3. Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014; 384:1376–88.
Article4. Smolle E, Taucher V, Haybaeck J. Malignant ascites in ovarian cancer and the role of targeted therapeutics. Anticancer Res. 2014; 34:1553–61.5. Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013; 3:256.
Article6. Choi YJ, Rhee JK, Hur SY, Kim MS, Lee SH, Chung YJ, et al. Intraindividual genomic heterogeneity of high-grade serous carcinoma of the ovary and clinical utility of ascitic cancer cells for mutation profiling. J Pathol. 2017; 241:57–66.
Article7. Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.
Article8. Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013; 497:108–12.
Article9. Husain H, Nykin D, Bui N, Quan D, Gomez G, Woodward B, et al. Cell-free DNA from ascites and pleural effusions: molecular insights into genomic aberrations and disease biology. Mol Cancer Ther. 2017; 16:948–55.
Article10. Zhou S, Xu B, Qi L, Zhu D, Liu B, Wei J. Next-generation sequencing reveals mutational accordance between cell-free DNA from plasma, malignant pleural effusion and ascites and directs targeted therapy in a gastric cancer patient. Cancer Biol Ther. 2019; 20:15–20.
Article11. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–90.
Article12. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2.13. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–9.
Article14. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–9.
Article15. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.
Article16. Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, et al. Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep. 2017; 7:4287.
Article17. Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016; 11:1–9.
Article18. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015; 31:2745–7.
Article19. Zhang Y, Cao L, Nguyen D, Lu H. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res. 2016; 5:650–63.
Article20. Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005; 11:2875–8.
Article21. Zeng M, Kwiatkowski NP, Zhang T, Nabet B, Xu M, Liang Y, et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. Elife. 2018; 7:e39030.
Article22. Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019; 393:1240–53.
Article23. Soletormos G, Duffy MJ, Othman Abu Hassan S, Verheijen RH, Tholander B, Bast RC Jr, et al. Clinical use of cancer biomarkers in epithelial ovarian cancer: updated huidelines from the European group on tumor markers. Int J Gynecol Cancer. 2016; 26:43–51.24. Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010; 116:1918–25.25. Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018; 56:186–97.
Article26. Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016; 13:e1002198.
Article27. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018; 359:926–30.
Article28. Chen X, Paranjape T, Stahlhut C, McVeigh T, Keane F, Nallur S, et al. Targeted resequencing of the microRNAome and 3'UTRome reveals functional germline DNA variants with altered prevalence in epithelial ovarian cancer. Oncogene. 2015; 34:2125–37.
Article29. Evans DGR, van Veen EM, Byers HJ, Wallace AJ, Ellingford JM, Beaman G, et al. A dominantly inherited 5' UTR variant causing methylation-associated silencing of BRCA1 as a cause of breast and ovarian cancer. Am J Hum Genet. 2018; 103:213–20.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Selective utilization of circulating tumor DNA testing enables disease monitoring in endometrial and ovarian carcinomas
- Clinical Circulating Tumor DNA Testing for Precision Oncology
- Loss of Heterozygosity using Microsatellite Marker in plasma of patients with ovarian cancer
- Significance of Interleukin-2(IL-2), Interleukin-6(IL-6), and Tumor Necrosis Factor-a(TNF-a) in the Ascites of Ovarian Cancer