Cancer Res Treat.
2020 Jan;52(1):74-84. 10.4143/crt.2019.062.
Fibroblast Growth Factor Receptor 1 (FGFR1) Amplification Detected by Droplet Digital Polymerase Chain Reaction (ddPCR) Is a Prognostic Factor in Colorectal Cancers
- Affiliations
-
- 1Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Laboratory of Epigenetics, Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- KMID: 2501203
- DOI: http://doi.org/10.4143/crt.2019.062
Abstract
- Purpose
The purpose of this study was to reveal the clinicopathological characteristics and prognostic implications associated with fibroblast growth factor receptor 1 (FGFR1) amplification in colorectal cancers (CRCs).
Materials and Methods
We measured the copy number of FGFR1 by droplet digital polymerase chain reaction (ddPCR), and analyzed the FGFR1 expression by immunohistochemistry, in 764 surgically resected CRCs (SNUH2007 dataset, 384 CRCs; SNUH Folfox dataset, 380 CRCs).
Results
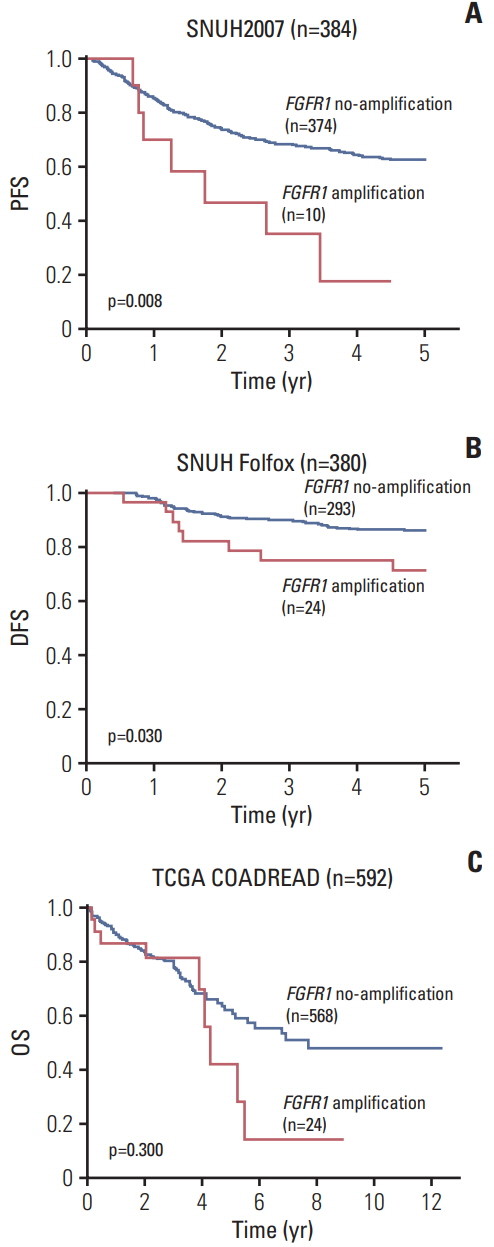
CRCs with ≥ 3.3 copies of the FGFR1 gene were classified as FGFR1 amplified. FGFR1 amplification was found in 10 of the 384 CRCs (2.6%) in the SNUH2007 dataset, and in 28 of the 380 CRCs (7.4%) in the SNUH Folfox dataset. In the SNUH2007 dataset, there was no association between the FGFR1 copy number status and sex, gross appearance, stage, or differentiation. High FGFR1 expression was associated with female sex and KRAS mutation. At the molecular level, FGFR1 amplification was mutually exclusive with BRAF mutation, microsatellite instability, and MLH1 methylation, in both SNUH2007 and SNUH Folfox datasets. Survival analysis revealed that FGFR1 amplification was associated with significantly worse clinical outcome compared with no FGFR1 amplification, in both SNUH2007 and SNUH Folfox datasets. Within the SNUH2007 dataset, CRC patients with high FGFR1 expression had an inferior progression-free survival compared with those with low FGFR1 expression. The FGFR inhibitor, PD173074, repressed the proliferation of a CRC cell line overexpressing FGFR1, but not of cells with FGFR1 amplification.
Conclusion
FGFR1 amplification measured by ddPCR can be a prognostic indicator of poor clinical outcome in patients with CRCs.
Keyword
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34.
Article2. Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019; 51:417–30.
Article3. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.4. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990; 61:759–67.
Article5. Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013; 45:1127–33.
Article6. Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014; 25:552–63.
Article7. Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013; 31:731–7.
Article8. Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget. 2015; 6:2562–72.
Article9. Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007; 9:R23.
Article10. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013; 31:3997–4013.
Article11. Fritzsche FR, Bode PK, Moch H, Kristiansen G, Varga Z, Bode B. Determination of the Her-2/neu gene amplification status in cytologic breast cancer specimens using automated silverenhanced in-situ hybridization (SISH). Am J Surg Pathol. 2010; 34:1180–5.
Article12. Yoo SB, Lee HJ, Park JO, Choe G, Chung DH, Seo JW, et al. Reliability of chromogenic in situ hybridization for epidermal growth factor receptor gene copy number detection in nonsmall-cell lung carcinomas: a comparison with fluorescence in situ hybridization study. Lung Cancer. 2010; 67:301–5.
Article13. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015; 61:79–88.
Article14. Goke F, Goke A, von Massenhausen A, Franzen A, Sharma R, Kirsten R, et al. Fibroblast growth factor receptor 1 as a putative therapy target in colorectal cancer. Digestion. 2013; 88:172–81.
Article15. Bae JM, Kim JH, Kwak Y, Lee DW, Cha Y, Wen X, et al. Distinct clinical outcomes of two CIMP-positive colorectal cancer subtypes based on a revised CIMP classification system. Br J Cancer. 2017; 116:1012–20.
Article16. Kinugasa H, Nouso K, Tanaka T, Miyahara K, Morimoto Y, Dohi C, et al. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Cancer. 2015; 112:1652–5.17. Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003; 349:247–57.
Article18. Yao TJ, Zhu JH, Peng DF, Cui Z, Zhang C, Lu PH. AZD-4547 exerts potent cytostatic and cytotoxic activities against fibroblast growth factor receptor (FGFR)-expressing colorectal cancer cells. Tumour Biol. 2015; 36:5641–8.
Article19. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–16.20. Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018; 33:721–35.21. Kwak Y, Nam SK, Seo AN, Kim DW, Kang SB, Kim WH, et al. Fibroblast growth factor receptor 1 gene copy number and mRNA expression in primary colorectal cancer and its clinicopathologic correlation. Pathobiology. 2015; 82:76–83.
Article22. Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010; 70:2085–94.23. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011; 83:8604–10.
Article24. Zhu Y, Lu D, Lira ME, Xu Q, Du Y, Xiong J, et al. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples. Exp Mol Pathol. 2016; 100:287–93.25. Koole K, Brunen D, van Kempen PM, Noorlag R, de Bree R, Lieftink C, et al. FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2016; 22:3884–93.26. Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–90.
Article27. Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009; 21:211–6.
Article28. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013; 13:772–87.
Article29. Paik PK, Shen R, Ferry D, Soria JC, Mathewson A, Kilgour E, et al. A phase 1b open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers: preliminary antitumor activity and pharmacodynamic data. J Clin Oncol. 2014; 32(15 Suppl):8035.
Article30. Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010; 2:62ra93.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Significance of HER-2/neu Oncogene Amplification Detected by Differential PCR in Advanced Human Ovarian Cancers
- A case of Pfeiffer syndrome with c833_834GC>TG (Cys278Leu) mutation in the FGFR2 gene
- Mutation Analysis in Fibroblast Growth Factor Receptor 3 Gene in Korean Children with Simple Craniosynostosis
- Association Study of Fibroblast Growth Factor 2 and Fibroblast Growth Factor Receptors Gene Polymorphism in Korean Ossification of the Posterior Longitudinal Ligament Patients
- Performance Evaluation of the QXDx BCR-ABL %IS Droplet Digital PCR Assay