J Lipid Atheroscler.
2020 Jan;9(1):153-161. 10.12997/jla.2020.9.1.153.
Select Macrophage Noncoding RNAs of Interest in Cardiovascular Disease
- Affiliations
-
- 1Department of Pathology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA. ebthorp@northwestern.edu
- KMID: 2470807
- DOI: http://doi.org/10.12997/jla.2020.9.1.153
Abstract
- Cardiovascular disease remains a leading cause of morbidity and mortality worldwide. Aspects of disease severity that are associated with heightened inflammation, such as during atherosclerosis or after myocardial infarction, are correlated with macrophage activation and macrophage polarization of the transcriptome and secretome. In this setting, non-coding RNAs (ncRNAs) may be as abundant as protein-coding genes and are increasingly recognized as significant modulators of macrophage gene expression and cytokine secretion, although the functions of most ncRNAs"”and in particular, long non-coding RNAs"”remain unknown. Herein, we discuss a subset of specific ncRNAs of interest in macrophages in atherosclerosis and during myocardial inflammation.
Keyword
MeSH Terms
Figure
Reference
-
1. Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation. 2019; 139:e56–e528.2. Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017; 70:1–25.3. Kinoshita M, Yokote K, Arai H, Iida M, Ishigaki Y, Ishibashi S, et al. Japan Atherosclerosis Society (JAS) guidelines for prevention of atherosclerotic cardiovascular diseases 2017. J Atheroscler Thromb. 2018; 25:846–984.
Article4. Bobryshev YV, Ivanova EA, Chistiakov DA, Nikiforov NG, Orekhov AN. Macrophages and their role in atherosclerosis: pathophysiology and transcriptome analysis. BioMed Res Int. 2016; 2016:9582430.
Article5. Otsuka F, Kramer MC, Woudstra P, Yahagi K, Ladich E, Finn AV, et al. Natural progression of atherosclerosis from pathologic intimal thickening to late fibroatheroma in human coronary arteries: a pathology study. Atherosclerosis. 2015; 241:772–782.
Article6. Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019; 20:29–39.
Article7. Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014; 111:16029–16034.
Article8. Wang L, Zhang YL, Lin QY, Liu Y, Guan XM, Ma XL, et al. CXCL1-CXCR2 axis mediates angiotensin II-induced cardiac hypertrophy and remodelling through regulation of monocyte infiltration. Eur Heart J. 2018; 39:1818–1831.
Article9. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014; 15:7–21.
Article10. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013; 152:1298–1307.
Article11. Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984; 81:1966–1970.
Article12. De Paepe B, Lefever S, Mestdagh P. How long noncoding RNAs enforce their will on mitochondrial activity: regulation of mitochondrial respiration, reactive oxygen species production, apoptosis, and metabolic reprogramming in cancer. Curr Genet. 2018; 64:163–172.
Article13. Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M, et al. Diabetes mellitus-induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler Thromb Vasc Biol. 2018; 38:1806–1820.
Article14. Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006; 25:6163–6169.
Article15. Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell. 2013; 49:298–309.
Article16. Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006; 38:758–769.
Article17. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012; 491:454–457.
Article18. Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016; 12:360–373.
Article19. Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187.
Article20. Ding M, Liu Y, Liao X, Zhan H, Liu Y, Huang W. Enhancer RNAs (eRNAs): new insights into gene transcription and disease treatment. J Cancer. 2018; 9:2334–2340.
Article21. Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018; 19:143–157.
Article22. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013; 152:570–583.
Article23. Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. 2014; 15:460.
Article24. Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics. 2011; 43:557–565.
Article25. O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016; 16:553–565.26. Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, Jepsen S, et al. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10 . Hum Mol Genet. 2013; 22:4516–4527.
Article27. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016; 7:12429.
Article28. Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2017; 58:470–480.
Article29. Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011; 470:264–268.
Article30. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, et al. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016; 13:98–108.
Article31. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011; 30:1956–1962.
Article32. Dutta P, Sager HB, Stengel KR, Naxerova K, Courties G, Saez B, et al. Myocardial infarction activates CCR2+ hematopoietic stem and progenitor cells. Cell Stem Cell. 2015; 16:477–487.
Article33. Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a . Nature. 2006; 443:421–426.
Article34. Li H, Han S, Sun Q, Yao Y, Li S, Yuan C, et al. Long non-coding RNA CDKN2B-AS1 reduces inflammatory response and promotes cholesterol efflux in atherosclerosis by inhibiting ADAM10 expression. Aging (Albany NY). 2019; 11:1695–1715.
Article35. Zhou X, Tao H, Cai Y, Cui L, Zhao B, Li K. Stage-dependent involvement of ADAM10 and its significance in epileptic seizures. J Cell Mol Med. 2019; 23:4494–4504.
Article36. van der Vorst EP, Weber C, Donners MM. A disintegrin and metalloproteases (ADAMs) in cardiovascular, metabolic and inflammatory diseases: aspects for theranostic approaches. Thromb Haemost. 2018; 118:1167–1175.
Article37. Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011; 25:444–448.38. Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010; 464:409–412.
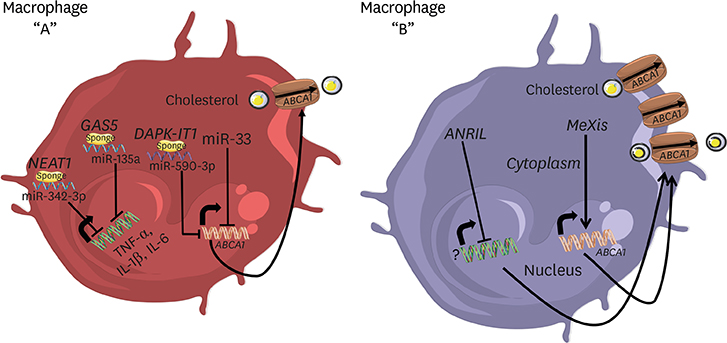
Article39. Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018; 24:304–312.
Article40. Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis . Nature. 2016; 534:124–128.
Article41. Zhang F, Wu L, Qian J, Qu B, Xia S, La T, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016; 75:96–104.
Article42. Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018; 17:90.
Article43. Yong W, Yu D, Jun Z, Yachen D, Weiwei W, Midie X, et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018; 9:861.
Article44. Gast M, Rauch BH, Haghikia A, Nakagawa S, Haas J, Stroux A, et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc Res. 2019; 115:1886–1906.
Article45. Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342-3p in human macrophages THP-1 cells. J Cell Physiol. 2019; 234:5319–5326.
Article46. Chen DD, Hui LL, Zhang XC, Chang Q. NEAT1 contributes to ox-LDL-induced inflammation and oxidative stress in macrophages through inhibiting miR-128. J Cell Biochem. 2018; 120:2493–2501.
Article47. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019; 10:1495.
Article48. Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010; 328:1570–1573.
Article49. Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, et al. Macrophage mitochondrial energy status regulates cholesterol efflux and is enhanced by anti-miR33 in atherosclerosis. Circ Res. 2015; 117:266–278.
Article50. Ouimet M, Ediriweera H, Afonso MS, Ramkhelawon B, Singaravelu R, Liao X, et al. microRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler Thromb Vasc Biol. 2017; 37:1058–1067.
Article51. Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011; 478:404–407.
Article52. Zhen Z, Ren S, Ji H, Ding X, Zou P, Lu J. The lncRNA DAPK-IT1 regulates cholesterol metabolism and inflammatory response in macrophages and promotes atherogenesis. Biochem Biophys Res Commun. 2019; 516:1234–1241.
Article53. Wang CH, Shi HH, Chen LH, Li XL, Cao GL, Hu XF. Identification of key lncRNAs associated with atherosclerosis progression based on public datasets. Front Genet. 2019; 10:123.
Article54. Yin Q, Wu A, Liu M. Plasma long non-coding RNA (lncRNA) GAS5 is a new biomarker for coronary artery disease. Med Sci Monit. 2017; 23:6042–6048.
Article55. Tao H, Zhang JG, Qin RH, Dai C, Shi P, Yang JJ, et al. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017; 386:11–18.
Article56. Ye J, Wang C, Wang D, Yuan H. LncRBA GSA5, up-regulated by ox-LDL, aggravates inflammatory response and MMP expression in THP-1 macrophages by acting like a sponge for miR-221. Exp Cell Res. 2018; 369:348–355.
Article57. Shen S, Zheng X, Zhu Z, Zhao S, Zhou Q, Song Z, et al. Silencing of GAS5 represses the malignant progression of atherosclerosis through upregulation of miR-135a. Biomed Pharmacother. 2019; 118:109302.
Article58. Ye ZM, Yang S, Xia YP, Hu RT, Chen S, Li BW, et al. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis. 2019; 10:138.
Article59. Thorp EB. Contrasting inflammation resolution during atherosclerosis and post myocardial infarction at the level of monocyte/macrophage phagocytic clearance. Front Immunol. 2012; 3:39.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Regulation Mechanism of Long Noncoding RNAs in Colon Cancer Development and Progression
- Potential Implications of Long Noncoding RNAs in Autoimmune Diseases
- Clinical significance of exosomal noncoding RNAs in hepatocellular carcinoma: a narrative review
- Noncoding RNAs as therapeutic targets in early stage diabetic kidney disease
- Role of Long Non-coding Ribonucleic Acid in Gastrointestinal Cancer