Healthc Inform Res.
2020 Jan;26(1):42-49. 10.4258/hir.2020.26.1.42.
Prediction of Drug–Drug Interactions by Using Profile Fingerprint Vectors and Protein Similarities
- Affiliations
-
- 1Department of Computer Engineering, Bahcesehir University, Besiktas, Istanbul, Turkey.
- 2Department of Software Engineering, Bahcesehir University, Besiktas, Istanbul, Turkey. serkan.ayvaz@eng.bau.edu.tr
- KMID: 2469399
- DOI: http://doi.org/10.4258/hir.2020.26.1.42
Abstract
OBJECTIVES
Drug-drug interaction (DDI) is a vital problem that threatens people's health. However, the prediction of DDIs through in-vivo experiments is not only extremely costly but also difficult as many serious side effects are hard to detect in in-vivo and in-vitro settings. The aim of this study was to assess the effectiveness of similarity-based in-silico computational DDI prediction approaches and to provide a cost effective and scalable solution to predict potential DDIs.
METHODS
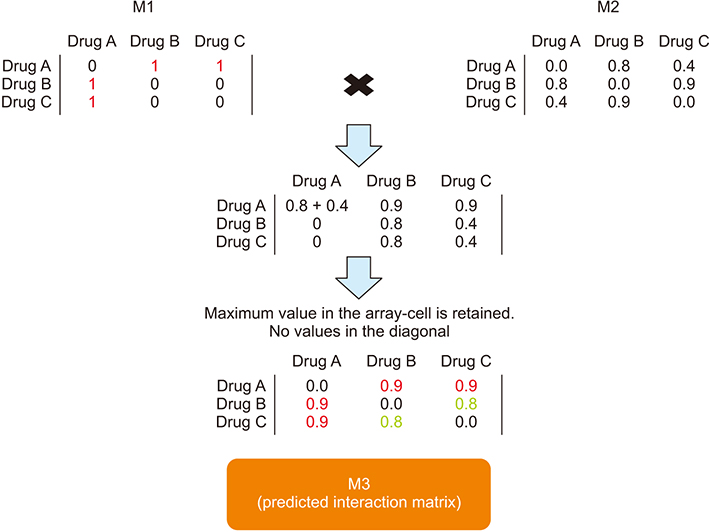
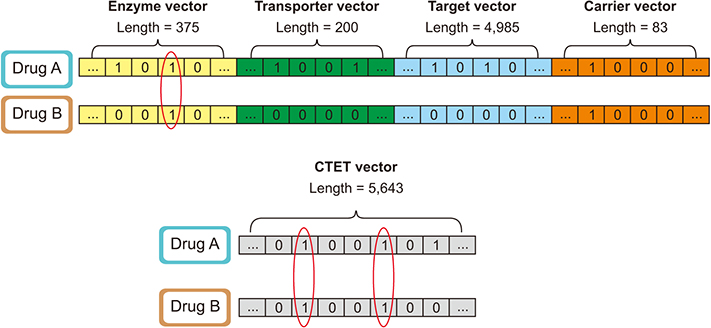
In this study, widely known similarity-based computational DDI prediction methods were utilized to discover novel potential DDIs. More specifically, known interactions, drug targets, adverse effects, and protein similarities of drug pairs were used to construct drug fingerprints for the prediction of DDIs.
RESULTS
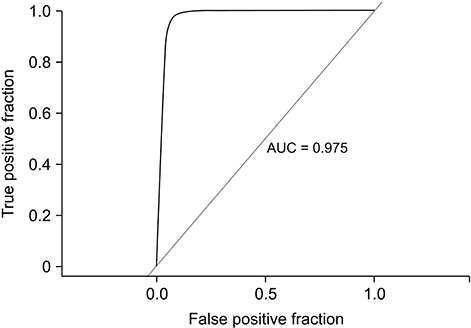
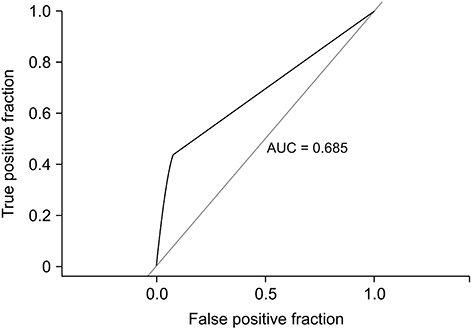
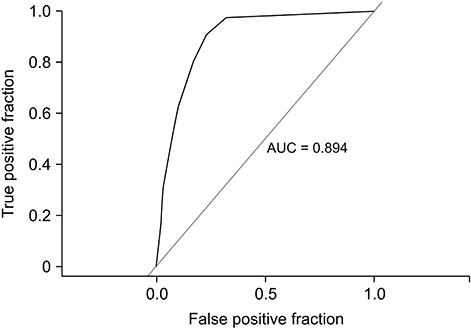
Using the drug interaction profile, our approach achieved an area under the curve (AUC) of 0.975 in the prediction of a potential DDI. The drug adverse effect profile and protein profile similarity-based methods resulted in AUC values of 0.685 and 0.895, respectively, in the prediction of DDIs.
CONCLUSIONS
In this study, we developed a computational approach to the prediction of potential drug interactions. The performance of the similarity-based computational methods was comparatively evaluated using a comprehensive real-world DDI dataset. The evaluations showed that the drug interaction profile information is a better predictor of DDIs compared to drug adverse effects and protein similarities among DDI pairs.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Estimating the Optimal Dexketoprofen Pharmaceutical Formulation with Machine Learning Methods and Statistical Approaches
Atakan Başkor, Yağmur Pirinçci Tok, Burcu Mesut, Yıldız Özsoy, Tamer Uçar
Healthc Inform Res. 2021;27(4):279-286. doi: 10.4258/hir.2021.27.4.279.
Reference
-
1. Vilar S, Uriarte E, Santana L, Lorberbaum T, Hripcsak G, Friedman C, et al. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nat Protoc. 2014; 9(9):2147–2163.
Article2. US Food and Drug Administration [Internet]. White Oak (MD): Food and Drug Administration;c2019. cited at 2020 Jan 19. Available from: http://www.fda.gov/.3. Cheng F, Zhao Z. Machine learning-based prediction of drug-drug interactions by integrating drug phenotypic, therapeutic, chemical, and genomic properties. J Am Med Inform Assoc. 2014; 21(e2):e278–e286.
Article4. Fowler S, Zhang H. In vitro evaluation of reversible and irreversible cytochrome P450 inhibition: current status on methodologies and their utility for predicting drug-drug interactions. AAPS J. 2008; 10(2):410–424.
Article5. Jonker DM, Visser SA, van der Graaf PH, Voskuyl RA, Danhof M. Towards a mechanism-based analysis of pharmacodynamic drug-drug interactions in vivo. Pharmacol Ther. 2005; 106(1):1–18.
Article6. Ferdousi R, Safdari R, Omidi Y. Computational prediction of drug-drug interactions based on drugs functional similarities. J Biomed Inform. 2017; 70:54–64.
Article7. Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Datadriven prediction of drug effects and interactions. Sci Transl Med. 2012; 4(125):125ra31.
Article8. Vilar S, Uriarte E, Santana L, Tatonetti NP, Friedman C. Detection of drug-drug interactions by modeling interaction profile fingerprints. PLoS One. 2013; 8(3):e58321.
Article9. Zhang W, Zou H, Luo L, Liu Q, Wu W, Xiao W. Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing. 2016; 173:979–987.
Article10. Ayvaz S, Horn J, Hassanzadeh O, Zhu Q, Stan J, Tatonetti NP, et al. Toward a complete dataset of drug-drug interaction information from publicly available sources. J Biomed Inform. 2015; 55:206–217.
Article11. Noor A, Assiri A, Ayvaz S, Clark C, Dumontier M. Drug-drug interaction discovery and demystification using Semantic Web technologies. J Am Med Inform Assoc. 2017; 24(3):556–564.
Article12. Vilar S, Harpaz R, Uriarte E, Santana L, Rabadan R, Friedman C. Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc. 2012; 19(6):1066–1074.
Article13. Vilar S, Uriarte E, Santana L, Friedman C, Tatonetti NP. State of the art and development of a drug-drug interaction large scale predictor based on 3D pharmacophoric similarity. Curr Drug Metab. 2014; 15(5):490–501.
Article14. Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen XW, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012; 19(e1):e28–e35.
Article15. Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inform Assoc. 2012; 19(1):79–85.
Article16. Harpaz R, Haerian K, Chase HS, Friedman C. Statistical mining of potential drug interaction adverse effects in FDA's spontaneous reporting system. AMIA Annu Symp Proc. 2010; 2010:281–285.17. Ibrahim H, Saad A, Abdo A, Sharaf Eldin A. Mining association patterns of drug-interactions using post marketing FDA's spontaneous reporting data. J Biomed Inform. 2016; 60:294–308.
Article18. Hudelson MG, Ketkar NS, Holder LB, Carlson TJ, Peng CC, Waldher BJ, et al. High confidence predictions of drug-drug interactions: predicting affinities for cytochrome P450 2C9 with multiple computational methods. J Med Chem. 2008; 51(3):648–654.
Article19. Percha B, Garten Y, Altman RB. Discovery and explanation of drug-drug interactions via text mining. In : Altman RB, Dunker AK, Hunter L, Murrey TA, Klein TE, editors. Biocomputing 2012. Singapore: World Scientific Publishing;2011. p. 410–421.20. Tari L, Anwar S, Liang S, Cai J, Baral C. Discovering drug-drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010; 26(18):i547–i553.
Article21. Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA). Drug Saf. 1999; 20(2):109–117.
Article22. Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006; 34(Database issue):D668–D672.
Article23. Fligner MA, Verducci JS, Blower PE. A modification of the Jaccard–Tanimoto similarity index for diverse selection of chemical compounds using binary strings. Technometrics. 2002; 44(2):110–119.
Article24. Willett P. Similarity-based approaches to virtual screening. Biochem Soc Trans. 2003; 31(Pt 3):603–606.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antidepressants and Related Drug Interactions
- Basic Principles of Drug Interaction
- Inhibition of Cytochrome P450 Enzymes by Drugs—Molecular Basis and Practical Applications
- Drug-drug Interactions between Psychotropic Agents and Other Drugs in Physically Ill Patients: Experience of Consultation-liason in Korea University Hospital
- Drug-Drug Interactions: Mood Stabilizers and Anti-Anxiety Drugs