Cancer Res Treat.
2019 Jul;51(3):1117-1127. 10.4143/crt.2018.405.
Genetic Profiles Associated with Chemoresistance in Patient-Derived Xenograft Models of Ovarian Cancer
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Women’s Cancer Center, Yonsei Cancer Center, Institute of Women's Life Medical Science, Yonsei University College of Medicine, Seoul, Korea. san1@yuhs.ac, nahmej6@yuhs.ac
- KMID: 2454303
- DOI: http://doi.org/10.4143/crt.2018.405
Abstract
- PURPOSE
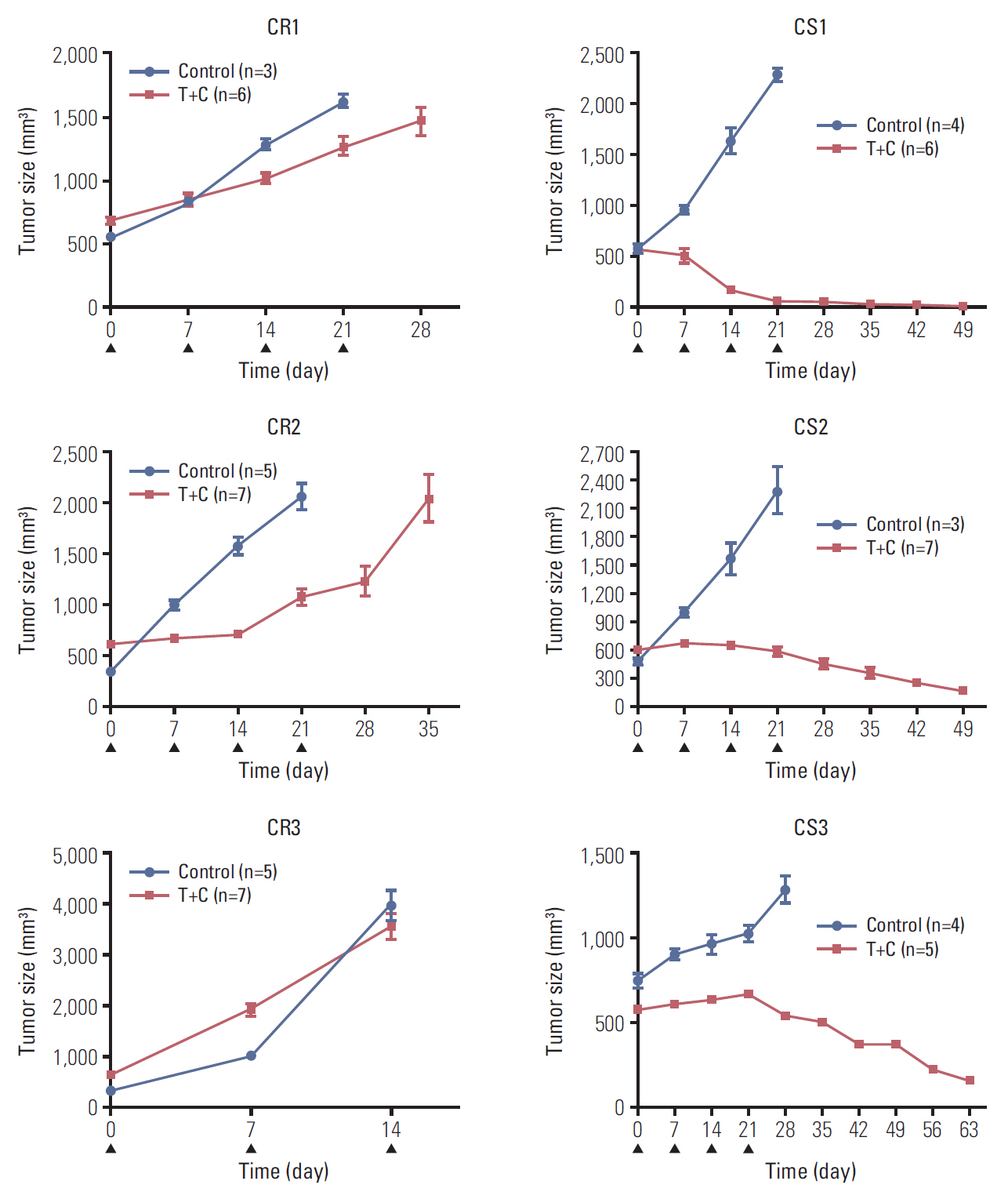
Recurrence and chemoresistance (CR) are the leading causes of death in patients with high-grade serous carcinoma (HGSC) of the ovary. The aim of this study was to identify genetic changes associated with CR mechanisms using a patient-derived xenograft (PDX) mouse model and genetic sequencing.
MATERIALS AND METHODS
To generate a CR HGSC PDX tumor, mice bearing subcutaneously implanted HGSC PDX tumors were treated with paclitaxel and carboplatin. We compared gene expression and mutations between chemosensitive (CS) and CR PDX tumors with whole exome and RNA sequencing and selected candidate genes. Correlations between candidate gene expression and clinicopathological variables were explored using the Cancer Genome Atlas (TCGA) database and the Human Protein Atlas (THPA).
RESULTS
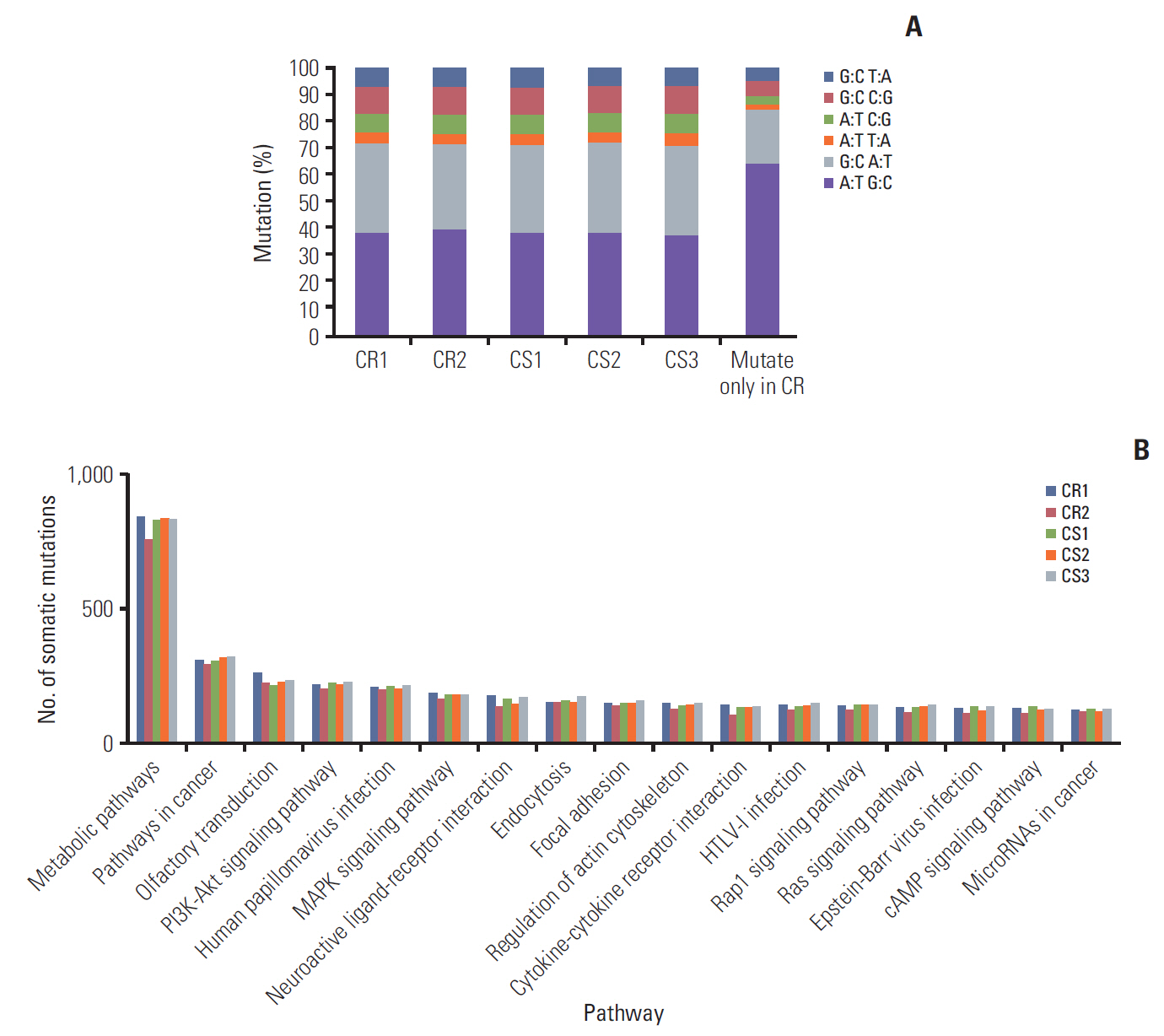
Three CR and four CS HGSC PDX tumor models were successfully established. RNA sequencing analysis of the PDX tumors revealed that 146 genes were significantly up-regulated and 54 genes down-regulated in the CR group compared with the CS group. Whole exome sequencing analysis showed 39 mutation sites were identified which only occurred in CR group. Differential expression of SAP25,HLA-DPA1, AKT3, and PIK3R5 genes and mutation of TMEM205 and POLR2A may have important functions in the progression of ovarian cancer chemoresistance. According to TCGA data analysis, patients with high HLA-DPA1 expression were more resistant to initial chemotherapy (p=0.030; odds ratio, 1.845).
CONCLUSION
We successfully established CR ovarian cancer PDX mouse models. PDX-based genetic profiling study could be used to select some candidate genes that could be targeted to overcome chemoresistance of ovarian cancer.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Romero I, Bast RC Jr. Minireview: human ovarian cancer: biology, current management, and paths to personalizing therapy. Endocrinology. 2012; 153:1593–602.
Article2. Holmes D. The problem with platinum. Nature. 2015; 527:S218–9.
Article3. Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007; 99:1441–54.
Article4. Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi MA, et al. The role of tumor microenvironment in chemoresistance: to survive, keep your enemies closer. Int J Mol Sci. 2017; 18:E1586.
Article5. Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015; 521:489–94.6. Bast RC Jr, Mills GB. Personalizing therapy for ovarian cancer: BRCAness and beyond. J Clin Oncol. 2010; 28:3545–8.
Article7. Dobbin ZC, Katre AA, Steg AD, Erickson BK, Shah MM, Alvarez RD, et al. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget. 2014; 5:8750–64.
Article8. Liu JF, Palakurthi S, Zeng Q, Zhou S, Ivanova E, Huang W, et al. Establishment of patient-derived tumor xenograft models of epithelial ovarian cancer for preclinical evaluation of novel therapeutics. Clin Cancer Res. 2017; 23:1263–73.
Article9. Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009; 15:1417–27.
Article10. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–60.
Article11. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a Map Reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.12. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21.
Article13. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28:511–5.
Article14. Rahman M, Hasan MR. Cancer metabolism and drug resistance. Metabolites. 2015; 5:571–600.
Article15. Ghazy AA, El-Etreby NM. Relevance of HLA-DP/DQ and ICAM-1 SNPs among ovarian cancer patients. Front Immunol. 2016; 7:202.
Article16. Kubler K, Arndt PF, Wardelmann E, Landwehr C, Krebs D, Kuhn W, et al. Genetic alterations of HLA-class II in ovarian cancer. Int J Cancer. 2008; 123:1350–6.17. Shiio Y, Rose DW, Aur R, Donohoe S, Aebersold R, Eisenman RN. Identification and characterization of SAP25, a novel component of the mSin3 corepressor complex. Mol Cell Biol. 2006; 26:1386–97.
Article18. Burris HA 3rd. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013; 71:829–42.
Article19. Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006; 441:424–30.
Article20. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–15.21. Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008; 27:5497–510.
Article22. Massacesi C, Di Tomaso E, Urban P, Germa C, Quadt C, Trandafir L, et al. PI3K inhibitors as new cancer therapeutics: implications for clinical trial design. Onco Targets Ther. 2016; 9:203–10.
Article23. Liby TA, Spyropoulos P, Buff Lindner H, Eldridge J, Beeson C, Hsu T, et al. Akt3 controls vascular endothelial growth factor secretion and angiogenesis in ovarian cancer cells. Int J Cancer. 2012; 130:532–43.
Article24. Burris HA, Siu LL, Infante JR, Wheler JJ, Kurkjian C, Opalinska J, et al. Safety, pharmacokinetics (PK), pharmacodynamics (PD), and clinical activity of the oral AKT inhibitor GSK2141795 (GSK795) in a phase I first-in-human study. J Clin Oncol. 2011; 29(15 Suppl):3003.
Article25. Shen DW, Ma J, Okabe M, Zhang G, Xia D, Gottesman MM. Elevated expression of TMEM205, a hypothetical membrane protein, is associated with cisplatin resistance. J Cell Physiol. 2010; 225:822–8.
Article26. Wang Y, Yin JY, Li XP, Chen J, Qian CY, Zheng Y, et al. The association of transporter genes polymorphisms and lung cancer chemotherapy response. PLoS One. 2014; 9:e91967.
Article27. Liu Y, Zhang X, Han C, Wan G, Huang X, Ivan C, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015; 520:697–701.28. Francavilla C, Lupia M, Tsafou K, Villa A, Kowalczyk K, Rakownikow Jersie-Christensen R, et al. Phosphoproteomics of primary cells reveals druggable kinase signatures in ovarian cancer. Cell Rep. 2017; 18:3242–56.
Article29. Yoo SS, Hong MJ, Lee JH, Choi JE, Lee SY, Lee J, et al. Association between polymorphisms in microRNA target sites and survival in early-stage non-small cell lung cancer. Thorac Cancer. 2017; 8:682–6.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of the Genetic Alterations between Primary Colorectal Cancers and Their Corresponding Patient-Derived Xenograft Tissues
- Chemoresistance in ovarian cancer: exploiting cancer stem cell metabolism
- Canine as a Comparative and Translational Model for Human Mammary Tumor
- Sex-dependent liver cancer xenograft models for predicting clinical data in the evaluation of anticancer drugs
- Establishing Patient-Derived Cancer Cell Cultures and Xenografts in Biliary Tract Cancer