Investig Clin Urol.
2019 May;60(3):148-155. 10.4111/icu.2019.60.3.148.
SETD2, GIGYF2, FGFR3, BCR, KMT2C, and TSC2 as candidate genes for differentiating multilocular cystic renal neoplasm of low malignant potential from clear cell renal cell carcinoma with cystic change
- Affiliations
-
- 1Department of Urology, Center for Prostate Cancer, Research Institute and National Cancer Center, Goyang, Korea. cjs5225@ncc.re.kr
- 2Department of Pathology, Center for Prostate Cancer, Research Institute and National Cancer Center, Goyang, Korea.
- KMID: 2444268
- DOI: http://doi.org/10.4111/icu.2019.60.3.148
Abstract
- PURPOSE
Multilocular cystic renal neoplasm of low malignant potential (MCRNLMP) and clear cell renal cell carcinoma with cystic change (MCRCC) have different prognoses despite similar histologic characteristics. The aim of this study was to identify differentially mutated genes in resected tumor specimens from patients diagnosed with MCRNLMP and MCRCC using a kidney cancer gene panel.
MATERIALS AND METHODS
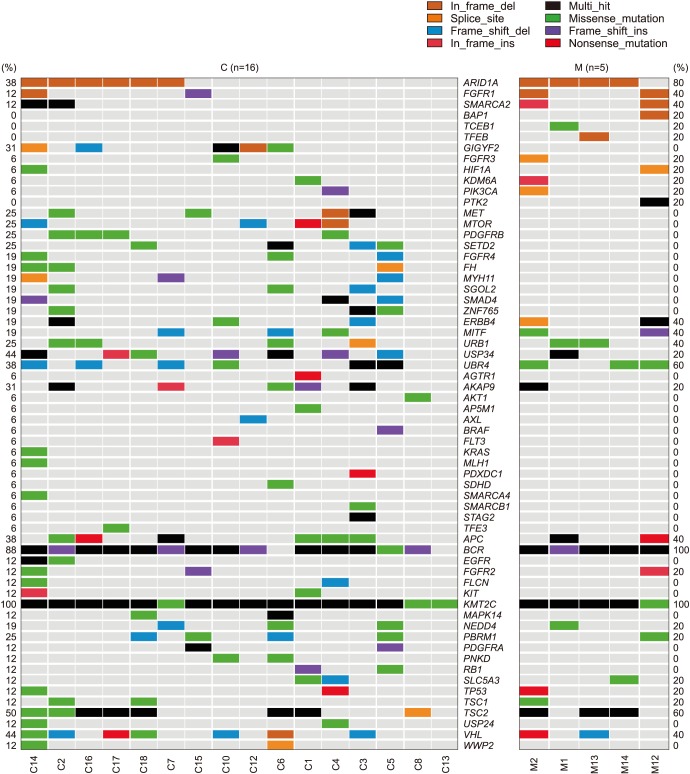
Between 2009 and 2016, 13 MCRNLMP and 17 MCRCC cases were selected. Tumor tissues from 5 MCRNLMP and 16 MCRCC cases were subjected to gene sequencing to detect mutations among 88 genes selected from a kidney cancer gene panel after quality control. Fisher's exact test was used to compare gene mutation profiles between the two diseases. Genes were considered to be positive for mutation according to the presence of an in-frame/frameshift deletion or insertion, missense/nonsense mutation, or multi-hit mutation.
RESULTS
During a median follow-up period of 66.2 months, there was only one case of MCRCC recurrence among all 30 patients. Target gene sequencing showed that 35 genes tended to be more frequently positive in either disease group, with six genes showing a significantly different frequency of mutation between the groups: GIGYF2 (odds ratio [OR], 5.735), FGFR3 (OR, 6.787), SETD2 (OR, 4.588), BCR (OR, 6.266), KMT2C (OR, 8.167), and TSC2 (OR, 4.474).
CONCLUSIONS
Six candidate genes showed significantly different mutation patterns between MCRNLMP and MCRCC, providing insight into their pathogenic mechanisms and differential prognoses.
Keyword
MeSH Terms
Figure
Reference
-
1. Kristiansen G, Delahunt B, Srigley JR, Lüders C, Lunkenheimer JM, Gevensleben H, et al. Vancouver classification of renal tumors: recommendations of the 2012 consensus conference of the International Society of Urological Pathology (ISUP). Pathologe. 2015; 36:310–316. PMID: 25398389.2. Srigley JR, Delahunt B, Eble JN, Egevad L, Epstein JI, Grignon D, et al. ISUP Renal Tumor Panel. The International Society of Urological Pathology (ISUP) Vancouver classification of renal neoplasia. Am J Surg Pathol. 2013; 37:1469–1489. PMID: 24025519.
Article3. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. 2016; 70:93–105. PMID: 26935559.
Article4. Montironi R, Lopez-Beltran A, Cheng L, Scarpelli M. Words of wisdom: re: multilocular cystic renal cell carcinoma with focus on clinical and pathobiological aspects. Eur Urol. 2013; 63:400–401. PMID: 23272732.5. Halat S, Eble JN, Grignon DJ, Lopez-Beltran A, Montironi R, Tan PH, et al. Multilocular cystic renal cell carcinoma is a subtype of clear cell renal cell carcinoma. Mod Pathol. 2010; 23:931–936. PMID: 20348877.
Article6. von Teichman A, Compérat E, Behnke S, Storz M, Moch H, Schraml P. VHL mutations and dysregulation of pVHL- and PTEN-controlled pathways in multilocular cystic renal cell carcinoma. Mod Pathol. 2011; 24:571–578. PMID: 21151099.
Article7. Kim SH, Park B, Joo J, Joung JY, Seo HK, Lee KH, et al. Retrospective analysis of 25 immunohistochemical tissue markers for differentiating multilocular cystic renal neoplasm of low malignant potential and multicystic renal cell carcinoma. Histol Histopathol. 2018; 33:589–596. PMID: 29292824.8. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–12.
Article9. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. q-bio.GN. 2013; arXiv:1303.3997.10. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. PMID: 19505943.
Article11. Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012; 3:35. PMID: 22435069.
Article12. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–219. PMID: 23396013.
Article13. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. PMID: 22728672.14. Yang J, Ding X, Sun X, Tsang SY, Xue H. SAMSVM: a tool for misalignment filtration of SAM-format sequences with support vector machine. J Bioinform Comput Biol. 2015; 13:1550025. PMID: 26419425.
Article15. Ajiro M, Nishidate T, Katagiri T, Nakamura Y. Critical involvement of RQCD1 in the EGFR-Akt pathway in mammary carcinogenesis. Int J Oncol. 2010; 37:1085–1093. PMID: 20878056.
Article16. Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG Jr. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003; 4:147–158. PMID: 12957289.
Article17. Ciccarese C, Brunelli M, Montironi R, Fiorentino M, Iacovelli R, Heng D, et al. The prospect of precision therapy for renal cell carcinoma. Cancer Treat Rev. 2016; 49:37–44. PMID: 27453294.
Article18. Durinck S, Stawiski EW, Pavía-Jiménez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015; 47:13–21. PMID: 25401301.
Article19. Fiorentino M, Gruppioni E, Massari F, Giunchi F, Altimari A, Ciccarese C, et al. Wide spectrum mutational analysis of metastatic renal cell cancer: a retrospective next generation sequencing approach. Oncotarget. 2017; 8:7328–7335. PMID: 27741505.20. Hsieh JJ, Le V, Cao D, Cheng EH, Creighton CJ. Genomic classifications of renal cell carcinoma: a critical step towards the future application of personalized kidney cancer care with panomics precision. J Pathol. 2018; 244:525–537. PMID: 29266437.
Article21. Li T, Chen J, Jiang Y, Ning X, Peng S, Wang J, et al. Multilocular cystic renal cell neoplasm of low malignant potential: a series of 76 cases. Clin Genitourin Cancer. 2016; 14:e553–e557. PMID: 27108004.
Article22. Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007; 18:460–466. PMID: 18023337.
Article23. Raspollini MR, Castiglione F, Martignoni G, Cheng L, Montironi R, Lopez-Beltran A. Unlike in clear cell renal cell carcinoma, KRAS is not mutated in multilocular cystic clear cell renal cell neoplasm of low potential. Virchows Arch. 2015; 467:687–693. PMID: 26438300.
Article24. Li J, Kluiver J, Osinga J, Westers H, van Werkhoven MB, Seelen MA, et al. Functional studies on primary tubular epithelial cells indicate a tumor suppressor role of SETD2 in clear cell renal cell carcinoma. Neoplasia. 2016; 18:339–346. PMID: 27292023.
Article25. Yang W, Ernst P. Distinct functions of histone H3, lysine 4 methyltransferases in normal and malignant hematopoiesis. Curr Opin Hematol. 2017; 24:322–328. PMID: 28375985.
Article26. Su D, Singer EA, Srinivasan R. Molecular pathways in renal cell carcinoma: recent advances in genetics and molecular biology. Curr Opin Oncol. 2015; 27:217–223. PMID: 25811348.27. Long C, Jian J, Li X, Wang G, Wang J. A comprehensive analysis of cancer-driving mutations and genes in kidney cancer. Oncol Lett. 2017; 13:2151–2160. PMID: 28454375.
Article28. Riazalhosseini Y, Lathrop M. Precision medicine from the renal cancer genome. Nat Rev Nephrol. 2016; 12:655–666. PMID: 27694978.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of multilocular cystic renal cell carcinoma mistaken for multilocular renal cyst
- A Case of Multilocular Cystic Renal Cell Carcinoma
- Multilocular Cystic Renal Cell Carcinoma: A case report
- Multilocular Cystic Renal Cell Carcinoma Treated with Wedge Resection
- Multilocular Cystic Renal Cell Carcinoma