World J Mens Health.
2019 May;37(2):148-156. 10.5534/wjmh.180071.
Epigenetics of Male Fertility: Effects on Assisted Reproductive Techniques
- Affiliations
-
- 1Department of Clinical and Experimental Medicine, University of Catania, Catania, Italy. sandrolavignera@unict.it
- KMID: 2443230
- DOI: http://doi.org/10.5534/wjmh.180071
Abstract
- During the last decades the study of male infertility and the introduction of the assisted reproductive techniques (ARTs) has allowed to understand that normal sperm parameters do not always predict fertilization. Sperm genetic components could play an important role in the early stages of embryonic development. Based on these acquisitions, several epigenetic investigations have been developed on spermatozoa, with the aim of understanding the multifactorial etiology of male infertility and of showing whether embryonic development may be influenced by sperm epigenetic abnormalities. This article reviews the possible epigenetic modifications of spermatozoa and their effects on male fertility, embryonic development and ART outcome. It focuses mainly on sperm DNA methylation, chromatin remodeling, histone modifications and RNAs.
MeSH Terms
Figure
Reference
-
1. Giacone F, Condorelli RA, Mongioì LM, Bullara V, La Vignera S, Calogero AE. In vitro effects of zinc, D-aspartic acid, and coenzyme-Q10 on sperm function. Endocrine. 2017; 56:408–415. PMID: 27422792.
Article2. Condorelli RA, La Vignera S, Mongioì LM, Vitale SG, Laganà AS, Cimino L, et al. Myo-inositol as a male fertility molecule: speed them up. Eur Rev Med Pharmacol Sci. 2017; 21:30–35.3. Cissen M, Wely MV, Scholten I, Mansell S, Bruin JP, Mol BW, et al. Measuring sperm DNA fragmentation and clinical outcomes of medically assisted reproduction: a systematic review and meta-analysis. PLoS One. 2016; 11:e0165125. PMID: 27832085.
Article4. Karaca MZ, Konac E, Yurteri B, Bozdag G, Sogutdelen E, Bilen CY. Association between methylenetetrahydrofolate reductase (MTHFR) gene promoter hypermethylation and the risk of idiopathic male infertility. Andrologia. 2017; 49. DOI: 10.1111/and.12698.
Article5. Jenkins TG, Aston KI, James ER, Carrell DT. Sperm epigenetics in the study of male fertility, offspring health, and potential clinical applications. Syst Biol Reprod Med. 2017; 63:69–76. PMID: 28301256.
Article6. Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014; 791:53–66. PMID: 23955672.
Article7. Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011; 727:62–71. PMID: 21540125.
Article8. Chen Q, Yan W, Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet. 2016; 17:733–743. PMID: 27694809.
Article9. Uysal F, Akkoyunlu G, Ozturk S. DNA methyltransferases exhibit dynamic expression during spermatogenesis. Reprod Biomed Online. 2016; 33:690–702. PMID: 27687053.
Article10. Gunes S, Agarwal A, Henkel R, Mahmutoglu AM, Sharma R, Esteves SC, et al. Association between promoter methylation of MLH1 and MSH2 and reactive oxygen species in oligozoospermic men: a pilot study. Andrologia. 2018; 50:DOI: 10.1111/and.12903.11. Owen CM, Segars JH Jr. Imprinting disorders and assisted reproductive technology. Semin Reprod Med. 2009; 27:417–428. PMID: 19711252.
Article12. Rahiminia T, Yazd EF, Fesahat F, Moein MR, Mirjalili AM, Talebi AR. Sperm chromatin and DNA integrity, methyltransferase mRNA levels, and global DNA methylation in oligoasthenoteratozoospermia. Clin Exp Reprod Med. 2018; 45:17–24. PMID: 29662821.
Article13. Olszewska M, Barciszewska MZ, Fraczek M, Huleyuk N, Chernykh VB, Zastavna D, et al. Global methylation status of sperm DNA in carriers of chromosome structural aberrations. Asian J Androl. 2017; 19:117–124. PMID: 26908061.
Article14. El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011; 5:60–69. PMID: 21293114.
Article15. Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006; 310:13–22. PMID: 16909904.
Article16. Cassidy FC, Charalambous M. Genomic imprinting, growth and maternal-fetal interactions. J Exp Biol. 2018; 221:pii: jeb164517.
Article17. Rai A, Cross JC. Development of the hemochorial maternal vascular spaces in the placenta through endothelial and vasculogenic mimicry. Dev Biol. 2014; 387:131–141. PMID: 24485853.
Article18. Santi D, De Vincentis S, Magnani E, Spaggiari G. Impairment of sperm DNA methylation in male infertility: a meta-analytic study. Andrology. 2017; 5:695–703. PMID: 28718528.
Article19. Camprubí C, Pladevall M, Grossmann M, Garrido N, Pons MC, Blanco J. Semen samples showing an increased rate of spermatozoa with imprinting errors have a negligible effect in the outcome of assisted reproduction techniques. Epigenetics. 2012; 7:1115–1124. PMID: 22885410.
Article20. Nanassy L, Carrell DT. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J Androl. 2011; 13:342–346. PMID: 21196940.
Article21. Nanassy L, Carrell DT. Abnormal methylation of the promoter of CREM is broadly associated with male factor infertility and poor sperm quality but is improved in sperm selected by density gradient centrifugation. Fertil Steril. 2011; 95:2310–2314. PMID: 21507395.22. Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, et al. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010; 25:2647–2654. PMID: 20685756.
Article23. Xu J, Zhang A, Zhang Z, Wang P, Qian Y, He L, et al. DNA methylation levels of imprinted and nonimprinted genes DMRs associated with defective human spermatozoa. Andrologia. 2016; 48:939–947. PMID: 26804237.
Article24. Sato A, Hiura H, Okae H, Miyauchi N, Abe Y, Utsunomiya T, et al. Assessing loss of imprint methylation in sperm from subfertile men using novel methylation polymerase chain reaction Luminex analysis. Fertil Steril. 2011; 95:129–134. 134.e1–134.e4. PMID: 20655520.
Article25. Laurentino S, Beygo J, Nordhoff V, Kliesch S, Wistuba J, Borgmann J, et al. Epigenetic germline mosaicism in infertile men. Hum Mol Genet. 2015; 24:1295–1304. PMID: 25336341.
Article26. Richardson ME, Bleiziffer A, Tüttelmann F, Gromoll J, Wilkinson MF. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet. 2014; 23:12–23. PMID: 23943794.
Article27. Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010; 94:1728–1733. PMID: 19880108.
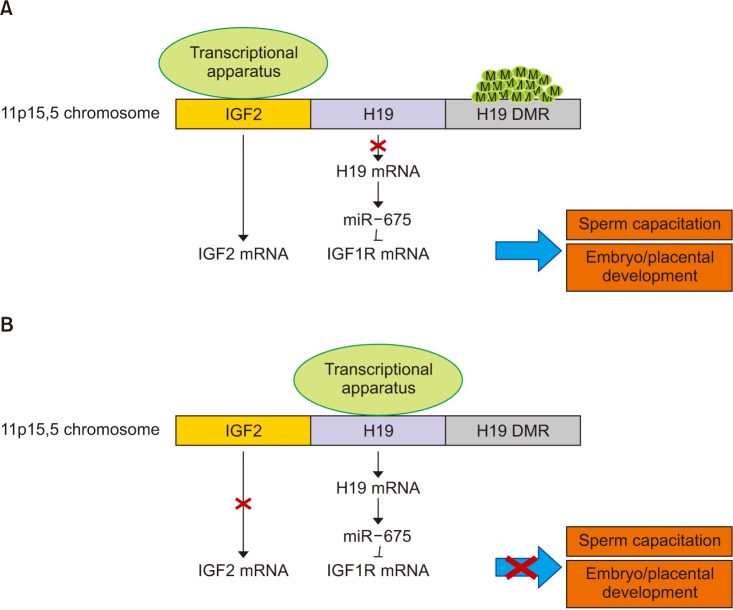
Article28. DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991; 64:849–859. PMID: 1997210.
Article29. Gao WL, Liu M, Yang Y, Yang H, Liao Q, Bai Y, et al. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal modulator 1 (NOMO1). RNA Biol. 2012; 9:1002–1010. PMID: 22832245.
Article30. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012; 14:659–665. PMID: 22684254.
Article31. Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993; 75:73–82. PMID: 8402902.
Article32. Constância M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002; 417:945–948. PMID: 12087403.
Article33. Fowden AL. The insulin-like growth factors and feto-placental growth. Placenta. 2003; 24:803–812. PMID: 13129676.
Article34. Qiu Q, Basak A, Mbikay M, Tsang BK, Gruslin A. Role of pro-IGF-II processing by proprotein convertase 4 in human placental development. Proc Natl Acad Sci U S A. 2005; 102:11047–11052. PMID: 16040806.
Article35. Arney KL. H19 and Igf2: enhancing the confusion? Trends Genet. 2003; 19:17–23. PMID: 12493244.36. Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004; 363:1700–1702. PMID: 15158633.
Article37. Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008; 14:67–74. PMID: 18178607.
Article38. Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010; 18:73–80. PMID: 19584898.
Article39. Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007; 16:2542–2551. PMID: 17636251.
Article40. Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T, et al. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta. 2015; 36:1100–1105. PMID: 26386650.
Article41. Wang J, Qi L, Huang S, Zhou T, Guo Y, Wang G, et al. Quantitative phosphoproteomics analysis reveals a key role of insulin growth factor 1 receptor (IGF1R) tyrosine kinase in human sperm capacitation. Mol Cell Proteomics. 2015; 14:1104–1112. PMID: 25693802.
Article42. Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012; 97:267–274. PMID: 22289286.
Article43. Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011; 141:21–36. PMID: 20876223.
Article44. Grassetti D, Paoli D, Gallo M, D'Ambrosio A, Lombardo F, Lenzi A, et al. Protamine-1 and -2 polymorphisms and gene expression in male infertility: an Italian study. J Endocrinol Invest. 2012; 35:882–888. PMID: 22104739.45. Nanassy L, Liu L, Griffin J, Carrell DT. The clinical utility of the protamine 1/protamine 2 ratio in sperm. Protein Pept Lett. 2011; 18:772–777. PMID: 21443494.
Article46. Lakpour N, Mahfouz RZ, Akhondi MM, Agarwal A, Kharrazi H, Zeraati H, et al. Relationship of seminal plasma antioxidants and serum male hormones with sperm chromatin status in male factor infertility. Syst Biol Reprod Med. 2012; 58:236–244. PMID: 22632096.
Article47. Utsuno H, Miyamoto T, Oka K, Shiozawa T. Morphological alterations in protamine-deficient spermatozoa. Hum Reprod. 2014; 29:2374–2381. PMID: 25190724.
Article48. Simon L, Castillo J, Oliva R, Lewis SE. Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod Biomed Online. 2011; 23:724–734. PMID: 22036908.
Article49. Francis S, Yelumalai S, Jones C, Coward K. Aberrant protamine content in sperm and consequential implications for infertility treatment. Hum Fertil (Camb). 2014; 17:80–89. PMID: 24869677.
Article50. Castillo J, Simon L, de Mateo S, Lewis S, Oliva R. Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J Androl. 2011; 32:324–332. PMID: 20966423.
Article51. La Vignera S, Condorelli RA, Vicari E, Calogero AE. Negative effect of increased body weight on sperm conventional and nonconventional flow cytometric sperm parameters. J Androl. 2012; 33:53–58. PMID: 21273503.
Article52. Condorelli RA, La Vignera S, Giacone F, Iacoviello L, Vicari E, Mongioi' L, et al. In vitro effects of nicotine on sperm motility and bio-functional flow cytometry sperm parameters. Int J Immunopathol Pharmacol. 2013; 26:739–746. PMID: 24067470.
Article53. Condorelli RA, La Vignera S, Giacone F, Iacoviello L, Mongioì LM, Li Volti G, et al. Nicotine effects and receptor expression on human spermatozoa: possible neuroendocrine mechanism. Front Physiol. 2017; 8:177. PMID: 28400736.
Article54. La Vignera S, Condorelli R, D'Agata R, Vicari E, Calogero AE. Semen alterations and flow-citometry evaluation in patients with male accessory gland infections. J Endocrinol Invest. 2012; 35:219–223. PMID: 21946047.55. La Vignera S, Condorelli R, Vicari E, D'Agata R, Calogero AE. Effects of varicocelectomy on sperm DNA fragmentation, mitochondrial function, chromatin condensation, and apoptosis. J Androl. 2012; 33:389–396. PMID: 21636732.
Article56. Condorelli R, Calogero AE, La Vignera S. Relationship between testicular volume and conventional or nonconventional sperm parameters. Int J Endocrinol. 2013; 2013:145792. PMID: 24089610.
Article57. Marchiani S, Tamburrino L, Benini F, Fanfani L, Dolce R, Rastrelli G, et al. Chromatin protamination and catsper expression in spermatozoa predict clinical outcomes after assisted reproduction programs. Sci Rep. 2017; 7:15122. PMID: 29123209.
Article58. Usachenko SI, Bradbury EM. Histone-DNA contacts in structure/ function relationships of nucleosomes as revealed by crosslinking. Genetica. 1999; 106:103–115. PMID: 10710716.59. Fournier C, Labrune E, Lornage J, Soignon G, Giscard d'Estaing S, Guérin JF, et al. The impact of histones linked to sperm chromatin on embryo development and ART outcome. Andrology. 2018; 6:436–445. PMID: 29499098.
Article60. Dadoune JP. Spermatozoal RNAs: what about their functions? Microsc Res Tech. 2009; 72:536–551. PMID: 19283828.
Article61. Zhao Y, Li Q, Yao C, Wang Z, Zhou Y, Wang Y, et al. Characterization and quantification of mRNA transcripts in ejaculated spermatozoa of fertile men by serial analysis of gene expression. Hum Reprod. 2006; 21:1583–1590. PMID: 16501037.
Article62. Curry E, Safranski TJ, Pratt SL. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology. 2011; 76:1532–1539. PMID: 21872314.
Article63. Yan W, Morozumi K, Zhang J, Ro S, Park C, Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and MicroRNAs in the sperm nuclei. Biol Reprod. 2008; 78:896–902. PMID: 18256326.64. Erickson RP. Post-meiotic gene expression. Trends Genet. 1990; 6:264–269. PMID: 1978427.
Article65. Vibranovski MD, Chalopin DS, Lopes HF, Long M, Karr TL. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics. 2010; 186:431–433. PMID: 20610406.
Article66. Fischer BE, Wasbrough E, Meadows LA, Randlet O, Dorus S, Karr TL, et al. Conserved properties of Drosophila and human spermatozoal mRNA repertoires. Proc Biol Sci. 2012; 279:2636–2644. PMID: 22378807.67. Bourc'his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010; 330:617–622. PMID: 21030645.68. Hosken DJ, Hodgson DJ. Why do sperm carry RNA? Relatedness, conflict, and control. Trends Ecol Evol. 2014; 29:451–455. PMID: 24916312.
Article69. Miller D, Ostermeier GC, Krawetz SA. The controversy, potential and roles of spermatozoal RNA. Trends Mol Med. 2005; 11:156–163. PMID: 15823753.
Article70. Boerke A, Dieleman SJ, Gadella BM. A possible role for sperm RNA in early embryo development. Theriogenology. 2007; 68(Suppl 1):S147–S155. PMID: 17583784.
Article71. Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 2016; 23:369–378. PMID: 26669700.
Article72. Xiao J, Wang X, Luo Y, Li XK, Li XW. Research progress in sRNAs and functional proteins in epididymosomes. Yi Chuan. 2018; 40:197–206. PMID: 29576543.73. Conine CC, Sun F, Song L, Rivera-Pérez JA, Rando OJ. Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev Cell. 2018; 46:470–480. PMID: 30057276.
Article74. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011; 12:246–258. PMID: 21427766.
Article